Abstract
Sensitivity to ethanol intoxication, propensity to drink ethanol and vulnerability to develop alcoholism are all influenced by genetic factors. Conversely, exposure to ethanol or subsequent withdrawal produce gene expression changes, which, in combination with environmental variables, may participate in the emergence of compulsive drinking and relapse. The present review offers an integrated perspective on brain gene expression profiling in rodent models of predisposition to differential ethanol sensitivity or consumption, in rats and mice subjected to acute or chronic ethanol exposure, as well as in human alcoholics. The functional categories over-represented among differentially expressed genes suggest that the transcriptional effects of chronic ethanol consumption contribute to the neuroplasticity and neurotoxicity characteristic of alcoholism. Importantly, ethanol produces distinct transcriptional changes within the different brain regions involved in intoxication, reinforcement and addiction. Special emphasis is put on recent profiling studies that have provided some insights into the molecular mechanisms potentially mediating genome-wide regulation of gene expression by ethanol. In particular, current evidence for a role of transcription factors, chromatin remodeling and microRNAs in coordinating the expression of large sets of genes in animals predisposed to excessive ethanol drinking or exposed to protracted abstinence, as well as in human alcoholics, is presented. Finally, studies that have compared ethanol with other drugs of abuse have highlighted common gene expression patterns that may play a central role in drug addiction. The availability of novel technologies and a focus on mechanistic approaches are shaping the future of ethanol transcriptomics.
Keywords: Ethanol, alcohol, gene expression, transcriptome, profiling, microarray, RNA-seq, brain
INTRODUCTION
Using Transcriptomics to Discover the Molecular Determinants of Alcoholism
Alcoholism is a chronic relapsing disorder characterized by compulsive intake of excessive amounts of ethanol, loss of control over drinking, emergence of a negative emotional state (e.g. depressed mood, anxiety, stress sensitivity) upon withdrawal, preoccupation with obtaining ethanol and narrowing of the behavioral repertoire at the expense of social, occupational and recreational activities, tolerance to the intoxicating effects of ethanol and continued drinking despite negative consequences (DSM-IV criteria for alcohol dependence). Alcoholism develops over the course of years and involves spiraling cycles of intoxication/withdrawal/craving [1]. Genetic factors account for more than 50% of the risk to develop alcoholism, according to a polygenic and epistatic scheme in which each individual gene only exerts a small influence and interact with other genes to impart vulnerability to alcoholism [2]. In individuals developing ethanol dependence, interplay between these predisposition genes, environmental factors and ethanol exposure in turn produces changes in gene expression, which are believed to contribute, together with epigenetic alterations and post-translational modifications, to the long-term allostatic changes in the activity of brain emotional systems that underlie alcoholism [3]. Exploration of ethanol-responsive genes in the brain is therefore expected to provide insights into the molecular mechanisms underlying compulsive drinking and relapse. These transcriptional changes are however anticipated to affect a broad set of genes, whose concerted regulation may orchestrate the complex behavioral outcome characterizing alcoholism. Gene expression profiling approaches are well suited to address this complexity because they enable hypothesis-free survey of transcriptional changes at a genome-wide scale.
A major challenge raised by genome-wide exploration of the transcriptome is to assign some biological significance to long lists of differentially expressed genes, but multiple tools are available to achieve that goal. First, clustering analysis can reveal patterns of transcriptional regulation shared by sets of genes, while gene promoter and 3’-untranslated region analysis can identify transcription factors and microRNAs (miRNAs) potentially coordinating the regulation of several transcripts at a time [4-7]. In addition, differential gene expression data sets can be overlaid onto databases integrating current knowledge on protein activity, functional interactions and higher-order relationships between proteins. Available bioinformatics resources include Gene Ontology (GO) classification (biological process, molecular function and cellular component, see [8]) and pathway-mining tools [9-12], which enable to identify networks of co-regulated genes whose products participate in the same biological function or signaling cascade. The use of genome-wide transcriptional profiling, associated with appropriate functional analysis, can therefore contribute to the identification of missing links in the sequence of molecular events leading to alcoholism [13].
Genome-wide analysis of ethanol-responsive genes has been the subject of intense research during the past decade. Microarray analysis of cultured neural cells (SH-SY5Y neuroblastoma cells) showed that ethanol exposure altered the expression of genes involved in cyclic AMP (cAMP)/protein kinase A (PKA) signaling and norepinephrine production, as well as in oxidative stress and protein synthesis [14, 15]. In the nematode, ethanol exposure rapidly induced a cellular stress response (heat shock protein genes) [16]. Gene profiling in the fruit fly also identified ethanol-responsive gene sets associated with stress response, along with olfaction, metabolism, transcription and signal transduction [17]. The present review focuses on whole-genome expression profiling in rodent genetic models of differential ethanol sensitivity or consumption, in rodents acutely or chronically exposed to ethanol and in human alcoholics. Importantly, rodent models and postmortem brain tissue have their own limitations and provide complementary biological information. While no animal paradigm can fully replicate the complexity of alcoholism, validated models enable the dissection of specific aspects of the syndrome with a tight control over experimental conditions and the possibility to use large numbers of isogenic subjects [18]. In contrast, the relevance of analyzing tissue from afflicted humans is unquestionable, but the genetic diversity of the subjects, the poor control over their history (and potential interference with co-morbid disorders), the limited availability of samples and variable integrity of RNA can potentially compromise the reliability and consistency of postmortem data [19].
An overview of all microarray studies conducted in rodent and human brain tissue is provided in Table 1. Earlier studies have been previously reviewed [20-24]. Most recent studies in the field of ethanol transcriptomics have improved our understanding of the genetic determinants of alcoholism by either of the following means:
Using rodent models of binge drinking and alcoholism with improved face and construct validity, such as chronic free choice drinking [25-29], induction of dependence [26, 30-34] and protracted abstinence [32-36].
Providing mechanistic insights into the regulation of gene expression by ethanol through the identification of transcription factors [37-40], epigenetic modifications [29, 35, 41, 42] and miRNAs [33, 34, 43] potentially coordinating changes in the expression of many transcripts at a time.
Integrating datasets to increase confidence in the identification of candidate genes (meta-analysis approach) and correlating gene expression levels with a relevant phenotype [29, 37-39, 44-47].
Comparing transcriptional patterns induced by ethanol with those induced by other drugs of abuse, in an effort to identify common factors influencing drug addiction [36, 42, 48].
Table 1.
Specifications of studies that have performed ethanol-related genome-wide gene expression profiling in the mammalian brain.
| Species/Strain or Line | Treatment | Brain Region(s) | Refs. |
|---|---|---|---|
| Models of Predisposition | |||
| Differential Ethanol Consumption/Preference | |||
| Alko, Alcohol vs Non-Alcohol (AA/ANA) rats selectively bred for high and low ethanol consumption | Naïve | NAc, Amg | [51] |
| FC | [100] | ||
| Inbred Preferring vs Non-Preferring (iP/iNP) rats selectively bred for high and low ethanol consumption | Naïve | Hipp | [57] |
| FC, Hipp, Amg, CPu, NAc | [56] | ||
| Rat congenital strains in which the iP chromosome 4 QTL region was introgressed onto iNP background | Naive | FC, Hipp, Amg, Str, NAc | [101] |
| 21 HXB/BXH recombinant inbred rat strains and progenitor strains with different levels of ethanol consumption | Naïve | Whole brain | [47] |
| C57Bl/6J vs DBA/2J mice, two inbred mouse strains with extreme (high vs low) levels of ethanol consumption | Saline injection | PFC, NAc, VTA | [54] |
| Meta-analysis of three selected mouse lines and six isogenic mouse strains with different levels of ethanol consumption | Naïve | Whole brain | [38] |
| High vs Low Alcohol Preference (HAP/LAP) mice and 20 BXD recombinant inbred mouse strains and progenitor strains with different levels of ethanol preference | Naïve | Whole brain | [39] |
| HAP/LAP mice, 30 BXD recombinant inbred strains and progenitor strains, and 20 inbred mouse strains with different levels of ethanol preference | Naïve | Whole brain | [46] |
| Differential Sensitivity to Acute Intoxication or Acute Functional Tolerance | |||
| Inbred Long-Sleep (ILS) and Short-Sleep (ISS) mice selectively bred for high or low sensitivity to ethanol-induced sedation | Naïve | Whole brain | [102] |
| Eight mouse inbred strains with different sensitivities to ethanol-induced locomotor activation | Naive | NAc, Cb | [103] |
| High vs Low Acute Functional Tolerance (HAFT/LAFT) mice selectively bred for high or low acute functional tolerance to ethanol-induced ataxia | Naïve | Whole brain | [40] |
| HAFT/LAFT mice and 26 BXD recombinant inbred strains and progenitor strains with different levels of ethanol acute functional tolerance | Naïve | Whole brain | [39] |
| HAFT/LAFT mice, 30 BXD recombinant inbred strains and progenitor strains, and 20 inbred mouse strains with different levels of ethanol acute functional tolerance | Naïve | Whole brain | [37] |
| Acute Exposure | |||
| DBA/2J mice | 4 g/kg ethanol (7 h) | Hipp | [53] |
| C57Bl/6J and DBA/2J mice | 6 g/kg ethanol (6 h) | Whole brain | [62] |
| C57Bl/6J and DBA/2J mice | 2 g/kg ethanol (4 h) | PFC, NAc, VTA | [54] |
| C57Bl/6J mice | 2.5 g/ethanol (2 h) | Midbrain | [104] |
| C57Bl/6J mice | 2 g/kg ethanol (1, 2 or 4 h) | Str | [48] |
| C57Bl/6J mice | 4 h access to 20% v/v ethanol (0 h) | OB, FC, Hipp, Str, VMB, Cb | [44] |
| Chronic Forced Exposure - Acute Withdrawal | |||
| Inhalation | |||
| C57Bl/6J and DBA/2J mice | 72 h ethanol vapor inhalation (7 h) | Hipp | [53] |
| Wistar rats | 2 weeks of chronic intermittent ethanol vapor inhalation (0 h) | mPFC, NAc, Amg | [31] |
| Withdrawal Seizure-Prone vs -Resistant (WSP/WSR) mice | 72-h ethanol vapor inhalation (8 h) | PFC | [63] |
| C57Bl/6J mice | 2 series of 4 cycles of intermittent ethanol vapor inhalation 1 week apart (0 or 8 h) | PFC, Hipp, NAc | [30] |
| Liquid Diet | |||
| Levis rats | 12% v/v ethanol liquid diet for 15 months | Hipp | [64] |
| Sprague-Dawley rats | 36% ethanol liquid diet for 21-28 weeks | NTS | [52] |
| Chronic Forced Exposure - Protracted Abstinence | |||
| Wistar rats | 7 weeks of chronic intermittent ethanol vapor inhalation (3 weeks) | mPFC, Amg | [32] |
| mPFC | [33, 34] | ||
| WSP/WSR mice | 72-h ethanol vapor inhalation (3 weeks) | PFC | [35] |
| Chronic Free-Choice Drinking | |||
| Meta-analysis of iP/iNP rats and postmortem human tissue gene expression, along with human genetic linkage data | Naïve, 10 weeks of continuous free-choice access to ethanol or eight 4-h sessions of intra-pVTA ethanol operant self-administration every other day | FC, Hipp, Amg, CPu, NAc | [45] |
| iP/iNP rats | 10 weeks of operant ethanol self-administration 1 h/day (24 h) | NAc, Amg | [28] |
| P rats | 8 weeks of free-choice continuous ethanol drinking (15 h) | NAc | [25] |
| 8 weeks of binge drinking: three daily 1-h sessions 5 days per week (1, 6 or 24 h) | NAcSh, CeA | [27] | |
| C57Bl/6N mice | Four cycles of four 18-h two-bottle choice (10% w/v ethanol/water) drinking sessions followed by four days of ethanol deprivation (6 days) | PFC, NAc, VMB | [29] |
| Chronic Ethanol Abuse | |||
| Human | Alcoholics (diagnosed by consumption, includes complicated cases) | sPFC | [43] |
| Human | Alcoholics (diagnosed by consumption) | Frontal and motor cortex | [55] |
| Human | Alcoholics (diagnosed by DSM-IV) with or without other psychiatric disorders | Temporal cortex | [65] |
| Human | Alcoholics (diagnosed by consumption) | Frontal and motor cortex | [59] |
| Human | Alcoholics (diagnosed by DSM-IV) | PFC | [75] |
| Human | Alcoholics (diagnosed by consumption and DSM-IV) | PFC | [58] |
| Human | Alcoholics (diagnosed by consumption) | sPFC | [61] |
| Human | Alcoholics (diagnosed by consumption) | sPFC | [60] |
| Human | Alcoholics vs cocaine abusers (DSM-IV) | Hipp | [42] |
| Human | Alcoholics (diagnosed by DSM-IV) | sPFC, BLA and CeA | [41] |
These studies have used rodent models of differential ethanol consumption or sensitivity, rodent models of acute or chronic forced exposure to ethanol followed by either acute withdrawal or protracted abstinence, rodent models of chronic free-choice drinking and human alcoholics. For each rodent model of ethanol exposure, the duration indicated between parentheses in the “Treatment” column corresponds to the time that elapsed between the termination of ethanol exposure and tissue sampling (this variable was not accessible for clinical studies).
Abbreviations for brain regions: OB, olfactory bulbs; FC, frontal cortex; PFC, prefrontal cortex; mPFC, medial prefrontal cortex; sPFC, superior frontal gyrus of the prefrontal cortex; Hipp, hippocampus; Amg, amygdala; CeA, central nucleus of the amygdala; Str, striatum; CPu, caudate-putamen; NAc, nucleus accumbens; NAcSh, nucleus accumbens shell; VMB, ventral midbrain; VTA, ventral tegmental area; NTS, nucleus of the tractus solitarius; Cb, cerebellum.
ETHANOL ALTERS GENE EXPRESSION ACROSS A WIDE RANGE OF BIOLOGICAL FUNCTIONS
Ethanol is a small molecule that interferes with numerous and diverse molecular targets, including neurotransmitter receptors and ion channels, and thereby modulates the activity of specific neural circuits to produce acute behavioral effects of ethanol such as dishinhibition, ataxia and sedation [49, 50]. Additional neurotransmitter and neuropeptide systems are then recruited during the acquisition and maintenance of ethanol drinking, and ultimately in the transition to ethanol dependence [1, 50]. Despite the variety of paradigms used to study the transcriptional effects of ethanol (see Table 1), a recurring conclusion from microarray studies is that ethanol affects a small proportion of the transcriptome (typically ~2%). A somewhat unexpected finding was that chronic binge drinking induces an even more limited set of transcriptional changes in the nucleus accumbens than chronic continuous drinking, which suggests that repeated episodes of intoxication and withdrawal lead to a tight regulation of gene expression, at least in this brain region [25, 27].
Table 2 gives an overview of the functional categories that have been identified across independent studies as enriched among differentially expressed genes. It is striking that all these categories repeatedly emerged from both preclinical and clinical studies and that in most cases they showed up across rodent models of predisposition, acute and chronic exposure. Genes related to neurotransmission, cell proliferation, neurite development and cytoskeleton speak to the structural and functional neuroplasticity induced by ethanol. Conversely, genes related to cell death, myelination and stress response are most relevant to the neurotoxic effects of ethanol. Importantly, some of these broad functional categories regroup a range of more specific molecular pathways or biological processes that may be differentially represented in the different paradigms of ethanol exposure.
Table 2.
Main functional categories identified by ethanol-related gene expression profiling.
| Functional Category | Rodent Models | Human Alcoholics | ||||
|---|---|---|---|---|---|---|
| Naïve (Models of Predisposition) | Acute Exposure | Chronic Forced Exposure | Chronic Free-Choice Drinking | Chronic Ethanol Abuse | ||
| Acute Withdrawal | Protracted Withdrawal | |||||
| Signal transduction | [38, 39, 51, 56, 57, 103] | [44, 48, 54, 62] | [30, 31, 52, 53] | [32, 33, 35] | [25, 27, 29] | [43, 55, 59, 60, 65, 75] |
| Neurotransmission and synaptic plasticity | [37, 40, 47, 56, 57, 100] | [54] | [52, 64] | [32-34] | [27-29] | [41, 60, 61] |
| Cell growth, proliferation, differentiation, adhesion, migration and neurite development | [46, 56, 57] | [48, 53, 54] | [53] | [25, 27-29, 45] | [43, 58-61] | |
| Cell death | [56, 57] | [48, 53, 62] | [52, 53, 63] | [35] | [27] | [43, 58-61] |
| Myelination | [56] | [54] | [34] | [43, 55, 59, 61, 75] | ||
| Transcription and epigenetic modifications | [38, 39, 56, 57, 104] | [44, 62] | [30, 63] | [35] | [25, 27, 29] | [41, 42, 58, 61, 75] |
| Stress response | [57] | [48, 62] | [52, 53, 63, 64] | [35] | [25, 29, 45] | [58-61, 65, 75] |
| Metabolism | [54, 57, 103] | [44] | [52] | [45] | [43, 58, 59, 61] | |
| Protein synthesis, trafficking and degradation | [54, 57] | [44] | [30, 53, 63] | [27] | [55, 59, 61, 65] | |
| Ion homeostasis, transport and binding | [56] | [44, 48, 54] | [52, 63] | [34, 35] | [28] | [60] |
| Cytoskeleton | [38, 51, 56] | [30] | [27] | [61] | ||
Specifications of rodent species/strain/line, treatment procedure, withdrawal duration, and brain region analyzed for each study can be found in Table 1. The listed functional categories intentionally correspond to broad, generic cellular functions or states, which emerged after collapsing more specific gene ontologies, molecular pathways or biological processes identified as enriched among differentially expressed genes in each referenced paper, in an effort to highlight convergent findings across rodent models and between preclinical and clinical data. Despite their breadth, these functional categories are not meant to be independent, mutually exclusive entities, and some of them show a significant degree of overlap (e.g., ion transport plays a critical role in neurotransmission and neurite development contributes to synaptic plasticity).
In particular, although the “Signal transduction” category has been consistently identified across studies, distinct signaling pathways were affected depending on the conditions of exposure. The mitogen-activated protein kinase (MAPK) cascade was identified in rodent models of differential ethanol consumption [38, 51] and following chronic ethanol exposure [30, 32, 52, 53]. Retinoic acid signaling was altered by acute and chronic ethanol treatment, as well as in alcoholics [53-55]. Rats with differential ethanol preference showed innate differences in small GTPase-mediated signal transduction, as did mice exposed to acute ethanol [48, 56]. Glucocorticoid signaling was altered following acute or chronic ethanol exposure [25, 54] and some of the transcriptional changes induced by acute ethanol exposure were reversed by an antagonist of the glucocorticoid receptor [48]. Chronic ethanol exposure, drinking or abuse affected the cAMP/PKA [32, 55], phosphoinositide 3-kinase [32, 53], calcium [25, 52, 55], and thyroid hormone [35, 55] signaling pathways. The notch and Janus kinase/signal transducers and activators of transcription (JAK/STAT) cascades were identified in individual studies using chronic ethanol exposure [30, 53]. In addition, a study focusing on a collection of hundred genes known to be regulated by chronic activation of the mu opioid receptor highlighted corticotropin releasing factor (CRF) and cdk5 signaling as being selectively targeted by excessive ethanol consumption in dependent mice, in contrast to moderate drinking or sole physical dependence [26]. Surprisingly, a CRF antagonist reversed some of the transcriptional changes induced by acute ethanol [48].
In the “Neurotransmission” category, glutamate transmission was identified in the whole brain of mice displaying differential ethanol acute functional tolerance, as well as in the prefrontal cortex of chronically drinking mice and human alcoholics [29, 37, 40, 41].
Two subdivisions of the “Cell growth” category, cell adhesion and migration, were altered in rodent models of differential ethanol consumption [46, 56, 57], in rodents subjected to chronic free-choice drinking [29, 45] and in human alcoholics [58-61].
Differential expression of genes related to chromatin remodeling (collapsed with those related to transcription in Table 2) was selectively identified in mouse models of chronic ethanol exposure [29, 35] and in brain tissue from human alcoholics [41, 42].
In most cases, genes in the “Stress response” category were related to oxidative stress and mitochondrial dysfunction, as found in models of predisposition [57] and following acute exposure [62], chronic exposure [52, 53, 63, 64], protracted abstinence [35], chronic free-choice drinking [25, 29], and chronic abuse in humans [58, 60, 65]. In few cases, they were related to immune response, following chronic exposure in mice [53], or chronic abuse in humans [59, 61].
In the “Protein” category, protein trafficking was identified in a rat model of differential ethanol preference [57], while ubiquitin-mediated protein degradation was altered following chronic ethanol inhalation [30, 53, 63]. Both processes however were affected in human alcoholics [55, 59, 61, 65]. Interestingly, an inhibitor of protein translation reversed some of the transcriptional changes induced by acute ethanol [48].
THE IMPORTANCE OF CIRCUITRY: ETHANOL EXPOSURE HAS DIFFERENT TRANSCRIPTIONAL EFFECTS DEPENDING ON THE BRAIN REGION
Gene profiling studies that have examined several brain regions at a time have showed that regional differences in gene expression were much larger than those induced by selective breeding for divergent ethanol preference [56] or by binge drinking [44]. Most importantly, such studies have revealed that ethanol-responsive gene expression patterns are brain region-specific, as summarized in Fig. (1) and as exemplified below.
Fig. (1).
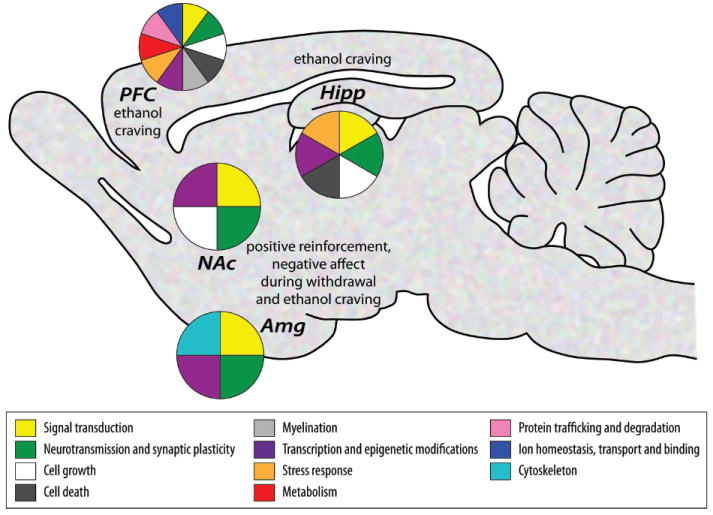
Brain scheme illustrating the regional specificity of functional categories that are enriched among genes differentially expressed in rodent predisposition models, rodent models of ethanol exposure and human alcoholics. Functional categories that were identified by at least two independent studies analyzing the same brain region are reported by a color-coded pie section. While “signal transduction”, “neurotransmission and synaptic plasticity” and “transcription and epigenetic modifications” emerged across all mesocorticolimbic structures, the other categories were selectively altered in individual regions. A vast majority of gene profiling studies performed in human alcoholics used postmortem prefrontal cortex (PFC) tissue and, as a result, most of the data generated in the nucleus accumbens (NAc), amygdala (Amg) and hippocampus (Hipp) arise from rodent models (see Table 1 for details). The role played by these four brain regions in the different stages of alcoholism are also highlighted. Amygdala was analyzed as a whole in some studies, while others have differentiated the central nucleus (implicated in acute reinforcement, emotional dysfunction in dependent subjects and stress-induced reinstatement) and the basolateral nucleus (implicated in cue conditioning, and, as such, in ethanol craving and relapse). Likewise, the nucleus accumbens is partitioned into a shell (contributing to the positive reinforcing effects of ethanol as well as to the negative emotional states associated with withdrawal), which was selectively sampled in some studies, and a core (involved in conditioned reinforcement). Note that other relevant brain regions (such as the ventral tegmental area, dorsal striatum or cerebellum) have been analyzed by a limited number of studies, which did not allow for the representation of convergent findings on this scheme.
A first example comes from a recent study that capitalized on a single episode of binge drinking to identify transcriptional changes correlated with blood alcohol levels [44]. C57Bl/6J mice were offered access to a bottle of water or 20% v/v ethanol for 4 h during the dark cycle, and blood and brain samples were obtained immediately at the end of the voluntary drinking session. Interestingly, the biological processes that were most populated with genes correlated with blood alcohol levels were brain region-specific: signal transduction in the cerebellum, ion transport in the hippocampus and ventral midbrain, gene expression in the olfactory bulb, metabolism in the frontal cortex, and transport/establishment of localization in the striatum. In the striatum, an ethanol-responsive module was enriched in genes known to be expressed in medium spiny neurons, indicating that this neuronal population is a sensitive target for acute ethanol-induced plasticity.
Discrepancies between brain regions also arose from models of chronic ethanol exposure. The effect of two series of four cycles of intermittent ethanol vapor inhalation one week apart, a paradigm known to induce physical dependence [66] and ethanol self-administration escalation [67], was analyzed in the prefrontal cortex, hippocampus and nucleus accumbens of C57Bl/6J mice [30]. Transcriptional disruption was strikingly stronger in the prefrontal cortex than in other brain regions at the end of chronic ethanol exposure, as previously found in rats exposed to two weeks of chronic intermittent ethanol inhalation [31]. Functional clustering of the 284 genes regulated in the prefrontal cortex highlighted the Ras/MAPK and notch signaling pathways, as well as ubiquitination. The hippocampus, on the other hand, exhibited a much higher transcriptional responsiveness during acute withdrawal, and several of the 139 regulated genes were associated with mRNA processing and actin dynamics. Genes governing circadian rhythms were over-represented among the few genes that were affected in the nucleus accumbens under both conditions. Interestingly, while the nucleus accumbens of C57Bl/6J mice was more responsive than prefrontal cortex to an acute ethanol challenge [54], an opposite pattern was found following chronic intermittent exposure to ethanol both in Wistar rats [31] and in C57Bl/6J mice [30], suggesting that tolerance to transcriptional disruption develops in the nucleus accumbens over the course of ethanol exposure, while the prefrontal cortex gets recruited later on in the process of ethanol dependence.
Another example is provided by the transcriptional effects of long-term free-choice ethanol drinking. A recent study took advantage of the inherent inter-individual variability in the ethanol intake of inbred C57Bl/6N mice to explore the “non-genetic” determinants of ethanol drinking [29]. Mice were subjected to four cycles of four 18-h two-bottle choice (10% w/v ethanol/water) drinking sessions, followed by four days of ethanol deprivation, and sacrificed 6 days after the last drinking session. Genes whose expression level correlated with stabilized ethanol intake (889 in nucleus accumbens, 850 in prefrontal cortex and 559 in ventral midbrain) were associated with glutamate signaling, brain-derived neurotrophic factor (BDNF) signaling, synaptic vesicle function and epigenetic modifications. In the nucleus accumbens, there was a striking enrichment for genes involved in transcriptional repression through epigenetic modifications (histone deacetylation and DNA methylation), and in synaptic vesicle formation and recycling. In the prefrontal cortex, mitochondrial dysfunction and glutamate signaling were highlighted, while functional categories over-represented in the ventral midbrain suggested that cell migration may be affected.
The vast majority of clinical studies have profiled gene expression in postmortem frontal cortex tissue of human alcoholics (see Table 1), but concomitant analysis of the central and basolateral nuclei of the amygdala recently provided evidence for the regional specificity of the transcriptional changes produced by alcoholism [41]. Gene coexpression network analysis was conducted to identify modules of genes whose expression co-varies across samples. Global coexpression profiles of cell-type-specific genes suggested that chronic ethanol abuse induces microglial activation in all three brain regions and neuronal degeneration in the amygdala. Interestingly, there was a coordinated upregulation of a cluster of genes involved in glutamatergic neurotransmission selectively in the cortex.
Altogether, these studies illustrate that within a same subject ethanol exposure affects the expression of genes belonging to different functional categories in distinct brain regions. The above findings clearly call for more studies on individual brain regions to address transcriptional adaptations relevant to the particular role of the neural substrates mediating the different aspects of ethanol intoxication, consumption and addiction [68]. These regions include cerebellum and brainstem nuclei for the motor and autonomic effects of ethanol; ventral tegmental area, nucleus accumbens and central nucleus of the amygdala for the acute reinforcing effects of ethanol; central nucleus of the amygdala, bed nucleus of the stria terminalis, and nucleus accumbens shell (which altogether compose the “extended amygdala” [69, 70]) for negative affect during ethanol withdrawal; medial prefrontal cortex, nucleus accumbens core, ventral pallidum, dorsal striatum, basolateral amygdala, hippocampus and extended amygdala for ethanol craving. In addition, neuroimaging studies in humans have also shown that behavioral impairments characteristic of alcoholism, including cognitive deficits and emotional dysfunction, are mediated by a disruption of frontal lobe and cerebellar circuitry [71].
Another parameter that has clearly been overlooked is the influence of gender on the genome-wide transcriptional effects of ethanol, as only two studies (by the same laboratory) have directly compared males and females so far. These two studies have examined the transcriptional effects of acute withdrawal and protracted abstinence from chronic ethanol exposure in mice selectively bred for divergent ethanol withdrawal severity (Withdrawal Seizure -Prone [WSP] and -Resistant [WSR] mice). In the first case, gene expression was analyzed in the prefrontal cortex 8 h into withdrawal from 72-h ethanol vapor inhalation [63]. Interestingly, sexual dimorphism had a stronger influence on the transcriptional response to chronic ethanol than differential susceptibility to ethanol withdrawal. Genes associated with transcription were identified in both genders, but genes involved in cell death and nucleic acid binding were preferentially regulated in females, while genes related to proteolysis and calcium ion binding were more responsive to chronic ethanol in males. Notably, induction of oxidative stress response was selectively observed in females. These findings suggest an enhanced sensitivity of females to ethanol-induced neurotoxicity, which was confirmed by histological analysis. In the second case, gene expression was analyzed 3 weeks after withdrawal from 72-h ethanol vapor inhalation [35]. In contrast to acute withdrawal, the mouse line (WSP vs WSR) accounted for more transcriptional variation than gender during protracted abstinence, which indicated that sexual dimorphism of the transcriptional response to ethanol strongly depends on the exposure paradigm.
IDENTIFICATION OF TRANSCRIPTION FACTORS CO-REGULATING THE EXPRESSION OF PREDISPOSING GENES
The specific molecular mechanisms through which ethanol regulates gene expression have been elucidated for a small number of genes [72]. In particular, a consensus sequence named alcohol response element (ARE) was identified in the second exon of the gene encoding the α4 subunit of the GABAA receptor (Gabra4) and proved to be essential for up-regulation of Gabra4 expression by ethanol in cortical neurons [73]. Further analysis revealed that the ARE sequence is a binding site for the transcription factor heat shock factor 1 (Hsf1) and that ethanol induces the nuclear translocation of Hsf1. Hsf1 was also shown to mediate the up-regulation of synaptotagmin 1 (Syt1) expression by ethanol, and ethanol induction of a subset of genes encoding synaptic vesicle fusion proteins correlated with the presence of multiple ARE sequences in promoter and intronic regions [74]. Interestingly, a highly homologous sequence, coined ethanol and stress response element (ESRE), was identified in the promoter region of several genes acutely induced by ethanol in the nematode, including several members of the heat shock protein family [16]. Accordingly, molecular chaperones were also differentially expressed in two strains of mice with divergent ethanol preference and up-regulated in the prefrontal cortex of alcoholics [54, 75]. Systematic in silico analysis of transcription factor binding sites over-represented in the promoter region of differentially expressed genes has been used in an attempt to identify transcription factors potentially orchestrating the regulation of candidate genes for ethanol preference and acute functional tolerance.
Analysis of whole brain gene expression in two replicate lines of mice selectively bred for high and low ethanol preference (HAP/LAP) identified 249 differentially expressed transcripts [39]. A list of eight candidate genes was generated based on their inclusion both in an expression quantitative trait locus (eQTL), as determined by global gene expression levels in BXD recombinant inbred strains, and in a behavioral quantitative trait locus (bQTL) for ethanol preference, as previously published [76, 77]. Only two of these genes, however, had expression levels correlated with ethanol intake in BXD recombinant inbred strains. Further analysis of the candidate gene promoters identified 23 over-represented transcription factor binding sites. Among those, binding motifs for Foxk2 and Tcf1 were present in the promoter of seven of the eight candidate genes, while Nfkb1 and Smad3 were localized within bQTL for ethanol preference. None of these transcription factors, however, were differentially expressed between HAP and LAP mice. A meta-analysis of whole brain gene expression in three groups of mouse lines selected for high and low ethanol intake (High Short-Term Selection [STS]/Low STS mice which were selected over 3 generations from a C57Bl/6J × DBA/2J reciprocal intercross, and two replicate lines of HAP/LAP mice) and six isogenic mouse strains known to differ markedly in voluntary ethanol consumption (C57Bl/6J, DBA/2J, BALB/cJ, LP/NJ, FVB/NJ and C57Bl/6J × FVB/NJ F1) identified ~3,800 unique genes whose expression levels were correlated with drinking levels [38]. Transcription factor binding sites for Zfp143 were present in the promoter of 64 genes positively correlated with ethanol intake, while consensus sequences for the fork-head box transcription factor Foxa2 were found in the promoter of 146 negatively correlated genes. Importantly, the overrepresented transcription factors had the same pattern of expression as their target genes, i.e. Zfp143 was up-regulated and Foxa2 was down-regulated in mouse models of high ethanol consumption. Although no causal relationship was established, these findings indicate that Zfp143 and Foxa2 could potentially coordinate the expression of genes predisposing to ethanol attraction or rejection. It would be interesting to investigate whether manipulating the expression of these two transcription factors would alter the expression of their target genes, and ultimately ethanol drinking.
Acute functional tolerance develops within a single exposure to ethanol and has been suggested to be a predisposing factor for the development of ethanol dependence [78]. A series of three studies by the same laboratory analyzed whole brain gene expression analysis in two replicate mouse lines selected for high or low acute functional tolerance to the loss of motor coordination produced by ethanol (HAFT/LAFT). The first study identified 144 differentially expressed transcripts [40]. Six of these genes lay within a bQTL for acute functional tolerance [79]. Binding sites for the transcription factor Cebpa were present in the promoter of five of the six candidate genes. The second study also combined gene expression and behavioral data from 26 BXD recombinant inbred strains, as described above for ethanol preference [39]. Twenty-two of the 275 genes differentially expressed in HAFT and LAFT mice were located both within an eQTL and a bQTL, and eight of these 22 candidate genes had expression levels correlated with acute functional tolerance to the incoordinating effects of ethanol in BXD strains. Further analysis of the candidate gene promoters identified 15 over-represented transcription factor binding sites. Among those, binding motifs for Usf1 were present in the promoter of 14 of the 22 candidate genes, while Srebf1 was localized within a bQTL for ethanol acute functional tolerance. The last study also integrated gene expression and acute functional tolerance data from 20 inbred mouse strains, as well as 30 BXD recombinant inbred strains [37]. Eight genes fulfilled the filtering criteria for the selection of candidate genes, including overlap between eQTL and bQTL, correlation between expression level and acute functional tolerance, and high heritability. Consensus sequences for the transcription factors Elk1, Arnt-Ahr, Irf1, Creb1 and E2f1 were present in five to seven of the eight candidate genes. A major limitation to the biological significance of these findings, however, is that none of the transcription factors whose binding motif was over-represented in the candidate gene promoters was differentially expressed in the brain of LAFT and HAFT mice. Moreover, there was a particularly poor overlap between the lists of over-represented transcription factors generated by these three studies, although they were all based on gene expression profiling in the whole brain of HAFT and LAFT mice, thereby challenging the role these transcription factors may play as key coordinators of gene expression patterns predisposing to high vs low acute functional tolerance to ethanol.
It therefore appears that efforts should be continued in the bioinformatics analysis of differentially expressed genes to uncover sequence patterns that may mediate gene co-regulation. Importantly, it is possible that relevant motifs still need to be identified and that some regulatory elements may not be located in the promoter region [e.g., 73].
CHROMATIN REMODELING AS A MECHANISM OF GENE EXPRESSION REGULATION BY CHRONIC ETHANOL
Chromatin remodeling designates the restructuring of histone proteins and DNA through reversible covalent modifications (e.g. histone acetylation and methylation, methylation of CpG islands in DNA), which produces conformational changes in the chromatin (condensed vs relaxed) and thereby modulates the accessibility of DNA to transcription factors and RNA polymerases [80]. These epigenetic modifications therefore have the ability to regulate gene expression without altering the DNA primary sequence. The role of epigenetic mechanisms in alcoholism is becoming an intense area of investigation [81, 82]. Accordingly, recent gene expression profiling studies have pinpointed that markers of chromatin remodeling were regulated by chronic ethanol exposure, both in rodent models and in human alcoholics, as detailed below.
In a study comparing gene expression profiles in the prefrontal cortex of WSP and WSR mice subjected to 72-h ethanol vapor inhalation and sacrificed 3 weeks later, a large set of transcripts involved in histone acetylation and deacetylation was down-regulated in WSR mice, but up-regulated in WSP mice, suggesting that epigenetic modifications may partly underlie persistent gene regulation during protracted abstinence [35]. Enrichment for genes involved in transcriptional repression through epigenetic modifications (histone deacetylation and DNA methylation) was also detected in the nucleus accumbens of C57Bl/6N mice subjected to four cycles of four 18-h two-bottle choice (10% w/v ethanol/water) drinking sessions, followed by four days of ethanol deprivation, and sacrificed 6 days after the last drinking session [29].
In a study conducted in postmortem hippocampal tissue samples from alcoholics, reverse transcribed RNA and genomic DNA fragments immunoprecipitated with an antibody against histone H3 trimethylated at lysine 4 (H3K4me3, a marker of active transcription) were subjected to high-throughput sequencing (RNA-seq) and there was no overlap between loci of H3K4me3 and gene expression changes [42]. This suggests that histone H3 lysine 4 trimethylation does not play a critical role in mediating transcriptional response to chronic ethanol abuse in the hippocampus.
In contrast, gene expression profiling in the superior frontal cortex, and central and basolateral nuclei of the amygdala of human alcoholics provided several lines of evidence (transcriptional activation of endogenous retrovirus transposons, downregulation of DNMT1 and upregulation of ribosomal modules) indicating that chronic ethanol abuse induces global DNA hypomethylation, which was hypothesized to reflect a release of transcriptional repression [41]. Moreover, gene coexpression network analysis pinpointed significantly overlapping modules, including those with extreme GC content (GC-rich/GC-poor), and there was a highly significant positive correlation between the direction and magnitude of gene regulation by chronic ethanol abuse and average GC content across modules identified in the three brain regions, which could be partially attributed to increased H3K4 trimethylation. Finally, chronic ethanol abuse induced a global increase in H3K4 trimethylation and an upregulation of several genes encoding elements of the transcription corepressor complex. Altogether this study points to chromatin modifications as a potential mechanism driving the coordination of gene expression patterns in the alcoholic brain.
MIRNAS AS A MECHANISM OF POST-TRANSCRIPTIONAL REGULATION BY CHRONIC ETHANOL
miRNAs are small (~22 nucleotides) non-coding mRNAs that can bind to complementary sequences located in the 3’-untranslated region of target mRNAs and promote degradation of the mRNA or repress its translation, thereby leading to expression silencing [83]. Importantly, each miRNA can control hundreds of mRNAs and each mRNA can be targeted by multiple miRNAs. Moreover, multiple miRNAs can either cooperate or compete for the regulation of common mRNA targets [e.g., 84, 85]. Regulation of miRNA levels by ethanol therefore represents a potential mechanism to coordinate expression levels of ethanol-responsive gene sets. Evidence for miRNA-mediated regulation of gene expression by ethanol was first provided by ex vivo studies. In neurosphere cultures derived from fetal mouse cortex, ethanol was shown to cause up-regulation of Jag1, a notch receptor ligand, through the combined suppression of miR-335, miR-21 and miR-153 [85]. In cultured neurons exposed to ethanol, miR-9 up-regulation altered the relative expression of splice variants of the alpha subunit of the large conductance calcium- and voltage-activated potassium (BK) channel (Kcnma1), thereby favoring the expression of BK isoforms resistant to ethanol-induced potentiation [86]. Systematic investigation of genome-wide effects of ethanol on miRNA expression in the adult brain has recently provided novel insights into the regulation of protein-coding gene expression in the context of chronic ethanol exposure.
miRNA expression profiling was performed in the frontal cortex of alcoholics [87]. A total of 35 miRNAs were up-regulated in alcoholics and used for target prediction. Interestingly, predicted miRNA targets were significantly over-represented in the set of 217 genes previously identified as down-regulated in the same postmortem samples, and most putative targets were regulated by multiple miRNAs [61]. Conversely, there were 27 miRNAs whose predicted targets were over-represented among down-regulated transcripts. Many of the down-regulated transcripts were related to lipid biosynthesis and metabolism, cytoskeleton and cell cycle control. Interestingly, the down-regulated transcript targeted by the highest number of up-regulated miRNAs encodes Dicer, a key enzyme in the generation of mature miRNAs. The authors proposed that such a negative feedback loop could serve as a mechanism for tight regulation of global miRNA levels and maintenance of cellular homeostasis.
Two recent, still unpublished, studies have concomitantly examined expression levels of protein-coding genes and miRNAs in the medial prefrontal cortex of Wistar rats exposed to 7 weeks of chronic intermittent inhalation of ethanol vapors, and sacrificed 3 weeks into withdrawal. A first study used an oligonucleotide array, along with a small non-coding RNA array [34, and personal communication from J. Tapocik]. A total of 165 genes were differentially expressed during protracted abstinence, and over-represented functional categories included myelination, ion channels, synaptic transmission and synaptic plasticity. An integrative analysis of the small and whole transcriptome datasets identified 33 miRNAs targeting 89 genes that were regulated in opposite directions. In particular, miR-9 was hypothesized to down-regulate 10 target genes. A second study used high-throughput sequencing to expand the range of profiled transcripts [33, and personal communication from J. Tapocik]. A total of 783 mRNAs and 861 small RNAs were differentially expressed in post-dependent animals. Network analysis highlighted interconnected genes involved in neurotransmission and signal transduction, and miRNAs miR-133b, miR-292-5p and miR-200 were hypothesized to orchestrate the regulation of this network.
COMPARISON OF THE TRANSCRIPTIONAL EFFECTS OF ETHANOL AND OTHER DRUGS OF ABUSE
Although ethanol and other drugs of abuse, such as opioids and psychostimulants, initially hit different molecular targets, the neurobiological mechanisms mediating positive reinforcement and transition to addiction strikingly share common neural substrates across drugs [68]. A couple of recent studies have undertaken to compare the transcriptional effects of ethanol with those of other drugs of abuse, in an effort to differentiate expression patterns specific to a given class of drug and to highlight commonly regulated genes that could play a central role in drug addiction.
A first study compared the time-dependent effects of acute exposure to six drugs of abuse (nicotine, ethanol, morphine, heroin, cocaine and methamphetamine) on gene expression (analyzed 1, 2, 4 and 8 h after injection) in the striatum of C57Bl/6J mice [48]. Clustering analysis identified two main modules of drug-responsive genes. A first module was selectively induced by psychostimulants (cocaine and methamphetamine) at early time-points and opioids (morphine and heroin) at later time-points, and contained several genes associated with transcription regulation, protein phosphatase activity and circadian rhythms. Induction of these genes by cocaine could be blocked by pre-treatment with dopamine D1 receptor antagonist or by a MEK1/2 inhibitor. The second module was subdivided into three subsets of genes selectively induced 1 to 2 h, 2 to 4 h or 4 h after injection of ethanol, respectively. These subsets were enriched in transcripts involved in 1. Small GTPase-mediated signal transduction, apoptosis and cell cycle control; 2. Enzyme inhibitor activity, apoptosis and stress response; and 3. Magnesium ion binding and morphogenesis. Interestingly, morphine and heroin produced a similar time-dependent gene expression pattern within these three subsets. In addition, an inhibitor of protein translation blocked induction of the first two subsets of genes by ethanol or morphine, while induction of the third subset of genes by ethanol could be blocked by prior administration of a CRF or a glucorticoid receptor antagonist.
Another comparative study analyzed gene expression in the extended amygdala (pooled samples of bed nucleus of the stria terminalis and central nucleus of the amygdala) of C57Bl/6J mice subjected to chronic exposure to four drugs of abuse (morphine, nicotine, Δ9-tetrahydrocannabinol and ethanol) or protracted abstinence (4 weeks) [36]. This study focused on a selection of hundred genes previously identified by genome-wide analysis as being regulated upon chronic activation of the mu opioid receptor. Principal component analysis revealed that the four drugs of abuse regulated distinct sets of genes immediately after treatment, but converged toward a common transcriptional signature following protracted abstinence. Clustering analysis identified one set of genes that were similarly down-regulated by all drugs 4 weeks into withdrawal. These nine genes were mapped to a network centered on the Htt gene and are known to be expressed in GABAergic medium spiny neurons.
Finally, a comparison of transcriptional and epigenetic changes induced by chronic cocaine or ethanol abuse in humans was conducted in postmortem hippocampus using whole transcriptome sequencing [42]. A total of 394 and 48 genes were differentially expressed in cocaine addicts and alcoholics, respectively, compared to drug-free subjects. There were 29 genes in common, including several transcriptional regulators and a number of small nucleolar RNAs, which guide chemical modification of pre-RNA molecules. Mitochondrial inner membrane function, however, was uniquely disrupted in the hippocampus of cocaine addicts.
FUTURE PERSPECTIVES
Most of the studies to date in the field of ethanol transcriptomics have used cDNA or oligonucleotide arrays. Microarrays have incorporated an increasing number of probes over the years and the most advanced ones can now profile expression of more than 45,000 transcripts at a time. High-throughput sequencing of the whole transcriptome (dubbed RNA-seq or next-generation sequencing), however, is opening new avenues for the understanding of gene expression tuning by ethanol. First of all, RNA-seq quantifies expression levels more accurately and has a larger dynamic range than microarrays [88]. In addition, RNA-seq uncovers the existence of novel transcripts that are not referenced in databases and are therefore not interrogated by microarrays. For instance, in a recent study using RNA-seq to compare gene expression in the prefrontal cortex of alcoholics and matched controls, the number of reads that did not map to RefSeq sequences represented more than one third of mapped reads [89, and personal communication from R.D. Mayfield]. Moreover, RNA-seq reveals sequence variations (such as single nucleotide polymorphisms, SNPs) within the transcripts. In the future, it would be interesting to determine whether ethanol differentially regulates the expression of SNP variants, in particular for SNPs previously shown by genome-wide association studies to confer vulnerability to or protection against alcoholism [2]. Finally, reads spanning exon junctions also inform about alternative splicing patterns. RNA-seq can therefore be used to assess whether ethanol alters expression of specific splice variants of individual genes, as was previously shown for Kcnma1 [86]. Accordingly, preliminary evidence for differential splice behavior of 125 genes emerged from transcriptome sequencing in the prefrontal cortex of alcoholics [89, and personal communication from R.D. Mayfield].
The wealth of gene expression data accumulated during the past decade has generated long lists of candidate genes predisposing to excessive ethanol drinking or responding to ethanol exposure or withdrawal. The vast majority of these genes, however, are still awaiting functional validation. Numerous studies using genetically engineered mice and virus-mediated gene silencing or overexpression have assessed the influence of individual genes on ethanol self-administration [90-99]. The next challenge will be to hinder or replicate the coordinated changes in gene expression produced by ethanol dependence and examine the effect of this global transcriptional remodeling on ethanol drinking. Realization of this ambitious goal will necessitate identifying the molecular mechanisms orchestrating these coordinated changes. Potential “master switches” have emerged from recent studies in the form of transcription factors, miRNAs and chromatin-modifying enzymes, as detailed in sections above and summarized in Fig. (2). It will be critical to expand these mechanistic approaches in the future, in the hope of identifying novel targets whose pharmacological modulation could address the complexity of molecular adaptations associated with alcoholism and could ultimately be used for therapeutic purposes.
Fig. (2).
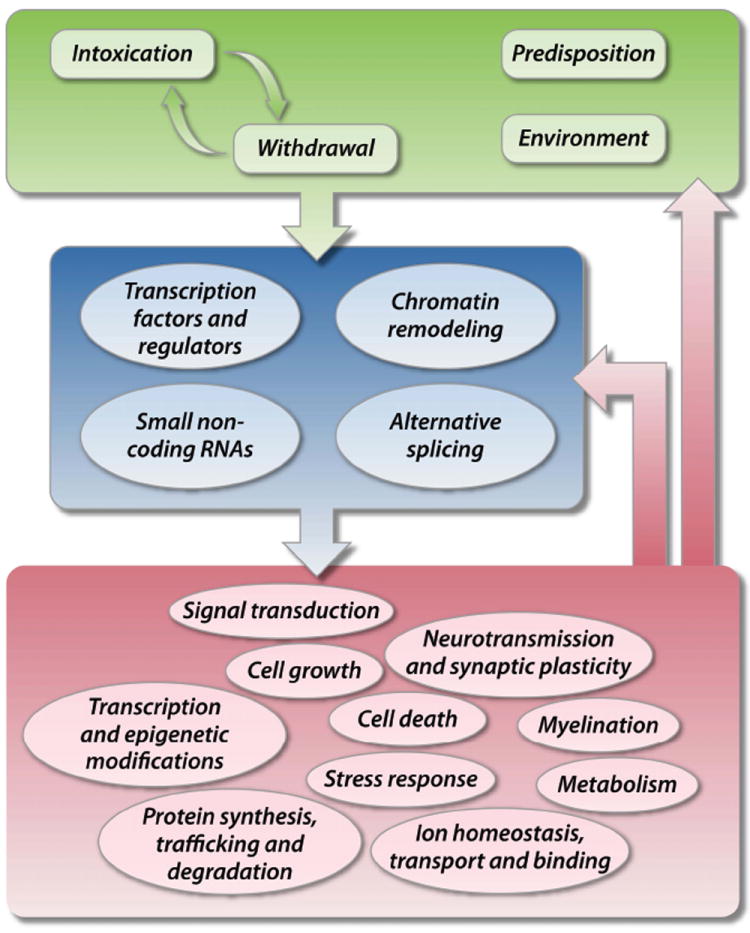
Schematic summary of molecular mechanisms hypothesized to drive the genome-wide regulation of gene expression in alcoholism. The green halo regroups potential causative factors for transcriptional changes in relation to alcoholism. Single nucleotide polymorphisms may influence the expression level of a given gene and the activity of its protein product, and thereby predispose an individual to increased vulnerability to or protection against alcoholism [2]. On the other hand, environmental variables, such as stressful life events or comorbid diseases, may also impact gene expression and thereby interact with the effects of ethanol [2]. The blue halo identifies molecular mechanisms that could mediate the global co-regulation of gene expression across the genome. Recent microarray and RNA-seq studies have pinpointed a potential role for transcription factors [37-40], epigenetic modifications [29, 35, 41, 42] and miRNAs [33, 34, 43] in the coordinated regulation of gene sets. In addition, RNA-seq is starting to uncover that alcoholism can affect the relative expression of different splice variants of a given gene [89]. The red halo highlights the functional categories that are enriched among genes differentially expressed in the brain of rodent predisposition models, rodent models of ethanol exposure and human alcoholics (see Table 2 for details). Changes in the expression level of genes encoding proteins involved in mRNA and small non-coding RNA processing, DNA methylation, histone acetylation and methylation can in turn contribute to accentuate, dampen or reorient the effects of the mechanisms identified in the blue halo. Moreover, changes in brain structure, connectivity and function resulting from the altered expression of genes involved in myelination, neurotransmission, signal transduction etc. are hypothesized to ultimately produce the behavioral impairments characteristic of alcoholism.
Acknowledgments
CC is funded by the National Institute on Alcohol Abuse and Alcoholism (Integrative Neuroscience Initiative on Alcoholism, U01 AA020913). This is manuscript #21699 from The Scripps Research Institute.
Footnotes
CONFLICT OF INTEREST
The author declares no conflict of interest.
References
- 1.Koob GF. Alcoholism: allostasis and beyond. Alcohol Clin Exp Res. 2003;27(2):232–43. doi: 10.1097/01.ALC.0000057122.36127.C2. [DOI] [PubMed] [Google Scholar]
- 2.Ducci F, Goldman D. Genetic approaches to addiction: genes and alcohol. Addiction. 2008;103(9):1414–28. doi: 10.1111/j.1360-0443.2008.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spanagel R. Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol Rev. 2009;89(2):649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- 4.Gonye GE, Chakravarthula P, Schwaber JS, Vadigepalli R. From promoter analysis to transcriptional regulatory network prediction using PAINT. Methods Mol Biol. 2007;408:49–68. doi: 10.1007/978-1-59745-547-3_4. [DOI] [PubMed] [Google Scholar]
- 5.Nugent R, Meila M. An overview of clustering applied to molecular biology. Methods Mol Biol. 2010;620:369–404. doi: 10.1007/978-1-60761-580-4_12. [DOI] [PubMed] [Google Scholar]
- 6.Matys V, Kel-Margoulis OV, Fricke E, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res. 2007;35(Web Server issue):W245–52. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 10.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas S, Bonchev D. A survey of current software for network analysis in molecular biology. Hum Genomics. 2010;4(5):353–60. doi: 10.1186/1479-7364-4-5-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werner T. Bioinformatics applications for pathway analysis of microarray data. Curr Opin Biotechnol. 2008;19(1):50–4. doi: 10.1016/j.copbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Farris SP, Miles MF. Ethanol modulation of gene networks: implications for alcoholism. Neurobiol Dis. 2012;45(1):115–21. doi: 10.1016/j.nbd.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman S, Miles MF. Identification of novel ethanol-sensitive genes by expression profiling. Pharmacol Ther. 2001;92(2-3):123–34. doi: 10.1016/s0163-7258(01)00163-2. [DOI] [PubMed] [Google Scholar]
- 15.Thibault C, Hassan S, Miles M. Using in vitro models for expression profiling studies on ethanol and drugs of abuse. Addict Biol. 2005;10(1):53–62. doi: 10.1080/13556210412331308949. [DOI] [PubMed] [Google Scholar]
- 16.Kwon JY, Hong M, Choi MS, et al. Ethanol-response genes and their regulation analyzed by a microarray and comparative genomic approach in the nematode Caenorhabditis elegans. Genomics. 2004;83(4):600–14. doi: 10.1016/j.ygeno.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Awofala AA. Genetic approaches to alcohol addiction: gene expression studies and recent candidates from Drosophila. Invert Neurosci. 2011;11(1):1–7. doi: 10.1007/s10158-010-0113-y. [DOI] [PubMed] [Google Scholar]
- 18.Markou A, Chiamulera C, Geyer MA, Tricklebank M, Steckler T. Removing obstacles in neuroscience drug discovery: the future path for animal models. Neuropsychopharmacology. 2009;34(1):74–89. doi: 10.1038/npp.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullumsmith RE, Meador-Woodruff JH. Novel approaches to the study of postmortem brain in psychiatric illness: old limitations and new challenges. Biol Psychiatry. 2011;69(2):127–33. doi: 10.1016/j.biopsych.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 20.Worst TJ, Vrana KE. Alcohol and gene expression in the central nervous system. Alcohol Alcohol. 2005;40(1):63–75. doi: 10.1093/alcalc/agh119. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman P, Tabakoff B. Gene expression in animals with different acute responses to ethanol. Addict Biol. 2005;10(1):63–9. doi: 10.1080/13556210412331308985. [DOI] [PubMed] [Google Scholar]
- 22.Flatscher-Bader T, van der Brug MP, Landis N, Hwang JW, Harrison E, Wilce PA. Comparative gene expression in brain regions of human alcoholics. Genes Brain Behav. 2006;5(Suppl 1):78–84. doi: 10.1111/j.1601-183X.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- 23.Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br J Pharmacol. 2008;154(2):275–87. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farris SP, Wolen AR, Miles MF. Using expression genetics to study the neurobiology of ethanol and alcoholism. Int Rev Neurobiol. 2010;91:95–128. doi: 10.1016/S0074-7742(10)91004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell RL, Kimpel MW, McClintick JN, et al. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav. 2009;94(1):131–47. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Contet C, Gardon O, Filliol D, Becker JA, Koob GF, Kieffer BL. Identification of genes regulated in the mouse extended amygdala by excessive ethanol drinking associated with dependence. Addict Biol. 2011;16(4):615–9. doi: 10.1111/j.1369-1600.2010.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride WJ, Kimpel MW, Schultz JA, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol. 2010;44(2):171–83. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodd ZA, Kimpel MW, Edenberg HJ, et al. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89(4):481–98. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolstenholme JT, Warner JA, Capparuccini MI, Archer KJ, Shelton KL, Miles MF. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS One. 2011;6(6):e21100. doi: 10.1371/journal.pone.0021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melendez RI, McGinty JF, Kalivas PW, Becker HC. Brain region-specific gene expression changes after chronic intermittent ethanol exposure and early withdrawal in C57BL/6J mice. Addict Biol. 2011 doi: 10.1111/j.1369-1600.2011.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Repunte-Canonigo V, Lutjens R, van der Stap LD, Sanna PP. Increased expression of protein kinase A inhibitor alpha (PKI-alpha) and decreased PKA-regulated genes in chronic intermittent alcohol exposure. Brain Res. 2007;1138:48–56. doi: 10.1016/j.brainres.2006.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rimondini R, Arlinde C, Sommer W, Heilig M. Long-lasting increase in voluntary ethanol consumption and transcriptional regulation in the rat brain after intermittent exposure to alcohol. Faseb J. 2002;16(1):27–35. doi: 10.1096/fj.01-0593com. [DOI] [PubMed] [Google Scholar]
- 33.Tapocik J, Barbier E, Schank J, et al. Small and whole transcriptome RNA sequencing identifies key regulation patterns in the medial prefrontal cortex of the alcohol dependent rat; ACNP 50th Annual Meeting; Waikoloa, HI. 2011. [Google Scholar]
- 34.Tapocik J, Sommer W, Schwandt M, Thorsell A, Heilig M. Gene expression profiling in the alcohol-dependent rat. Alcohol Clin Exp Res. 2011;35(s1):262A. [Google Scholar]
- 35.Hashimoto JG, Forquer MR, Tanchuck MA, Finn DA, Wiren KM. Importance of genetic background for risk of relapse shown in altered prefrontal cortex gene expression during abstinence following chronic alcohol intoxication. Neuroscience. 2011;173:57–75. doi: 10.1016/j.neuroscience.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Merrer J, Befort K, Gardon O, et al. Protracted abstinence from distinct drugs of abuse shows regulation of a common gene network. Addict Biol. 2012;17(1):1–12. doi: 10.1111/j.1369-1600.2011.00365.x. [DOI] [PubMed] [Google Scholar]
- 37.Hu W, Saba L, Kechris K, Bhave SV, Hoffman PL, Tabakoff B. Genomic insights into acute alcohol tolerance. J Pharmacol Exp Ther. 2008;326(3):792–800. doi: 10.1124/jpet.108.137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulligan MK, Ponomarev I, Hitzemann RJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103(16):6368–73. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saba L, Bhave SV, Grahame N, et al. Candidate genes and their regulatory elements: Alcohol preference and tolerance. Mamm Genome. 2006;17(6):669–88. doi: 10.1007/s00335-005-0190-0. [DOI] [PubMed] [Google Scholar]
- 40.Tabakoff B, Bhave SV, Hoffman PL. Selective breeding, quantitative trait locus analysis, and gene arrays identify candidate genes for complex drug-related behaviors. J Neurosci. 2003;23(11):4491–8. doi: 10.1523/JNEUROSCI.23-11-04491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32(5):1884–97. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci U S A. 2011;108(16):6626–31. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24(12):1873–82. [PubMed] [Google Scholar]
- 44.Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Adron Harris R, Ponomarev I. Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol Clin Exp Res. 2011;35(4):659–70. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodd ZA, Bertsch BA, Strother WN, et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2007;7(4):222–56. doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- 46.Tabakoff B, Saba L, Kechris K, et al. The genomic determinants of alcohol preference in mice. Mamm Genome. 2008;19(5):352–65. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tabakoff B, Saba L, Printz M, et al. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piechota M, Korostynski M, Solecki W, et al. The dissection of transcriptional modules regulated by various drugs of abuse in the mouse striatum. Genome Biol. 2010;11(5):R48. doi: 10.1186/gb-2010-11-5-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris RA, Trudell JR, Mihic SJ. Ethanol’s molecular targets. Sci Signal. 2008;1(28):re7. doi: 10.1126/scisignal.128re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. Br J Pharmacol. 2008;154(2):299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arlinde C, Sommer W, Bjork K, et al. A cluster of differentially expressed signal transduction genes identified by microarray analysis in a rat genetic model of alcoholism. Pharmacogenomics J. 2004;4(3):208–18. doi: 10.1038/sj.tpj.6500243. [DOI] [PubMed] [Google Scholar]
- 52.Covarrubias MY, Khan RL, Vadigepalli R, Hoek JB, Schwaber JS. Chronic alcohol exposure alters transcription broadly in a key integrative brain nucleus for homeostasis: the nucleus tractus solitarius. Physiol Genomics. 2005;24(1):45–58. doi: 10.1152/physiolgenomics.00184.2005. [DOI] [PubMed] [Google Scholar]
- 53.Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain Behav. 2002;1(1):35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- 54.Kerns RT, Ravindranathan A, Hassan S, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25(9):2255–66. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81(4):802–13. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- 56.Kimpel MW, Strother WN, McClintick JN, et al. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41(2):95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edenberg HJ, Strother WN, McClintick JN, et al. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4(1):20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- 58.Flatscher-Bader T, van der Brug M, Hwang JW, et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93(2):359–70. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- 59.Liu J, Lewohl JM, Dodd PR, Randall PK, Harris RA, Mayfield RD, et al. Gene expression profiling of individual cases reveals consistent transcriptional changes in alcoholic human brain. J Neurochem. 2004;90(5):1050–8. doi: 10.1111/j.1471-4159.2004.02570.x. [DOI] [PubMed] [Google Scholar]
- 60.Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcohol Clin Exp Res. 2007;31(9):1460–6. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Lewohl JM, Harris RA, et al. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31(7):1574–82. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- 62.Treadwell JA, Singh SM. Microarray analysis of mouse brain gene expression following acute ethanol treatment. Neurochem Res. 2004;29(2):357–69. doi: 10.1023/b:nere.0000013738.06437.a6. [DOI] [PubMed] [Google Scholar]
- 63.Hashimoto JG, Wiren KM. Neurotoxic consequences of chronic alcohol withdrawal: expression profiling reveals importance of gender over withdrawal severity. Neuropsychopharmacology. 2008;33(5):1084–96. doi: 10.1038/sj.npp.1301494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saito M, Smiley J, Toth R, Vadasz C. Microarray analysis of gene expression in rat hippocampus after chronic ethanol treatment. Neurochem Res. 2002;27(10):1221–9. doi: 10.1023/a:1020937728506. [DOI] [PubMed] [Google Scholar]
- 65.Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72(6):756–67. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- 66.Becker HC, Hale RL. Repeated episodes of ethanol withdrawal potentiate the severity of subsequent withdrawal seizures: an animal model of alcohol withdrawal “kindling”. Alcohol Clin Exp Res. 1993;17(1):94–8. doi: 10.1111/j.1530-0277.1993.tb00731.x. [DOI] [PubMed] [Google Scholar]
- 67.Becker HC, Lopez MF. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 2004;28(12):1829–38. doi: 10.1097/01.alc.0000149977.95306.3a. [DOI] [PubMed] [Google Scholar]
- 68.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35(1):217–38. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heimer L, Alheid G. Piecing together the puzzle of basal forebrain anatomy. In: Napier TC, Kalivas PW, Hanin I, editors. The Basal Forebrain: Anatomy to Function. New York: Plenum Press; 1991. pp. 1–42. [DOI] [PubMed] [Google Scholar]
- 70.Koob GF, Roberts AJ, Schulteis G, et al. Neurocircuitry targets in ethanol reward and dependence. Alcohol Clin Exp Res. 1998;22(1):3–9. [PubMed] [Google Scholar]
- 71.Sullivan EV, Pfefferbaum A. Neurocircuitry in alcoholism: a substrate of disruption and repair. Psychopharmacology (Berl) 2005;180(4):583–94. doi: 10.1007/s00213-005-2267-6. [DOI] [PubMed] [Google Scholar]
- 72.Pignataro L, Varodayan FP, Tannenholz LE, Harrison NL. The regulation of neuronal gene expression by alcohol. Pharmacol Ther. 2009;124(3):324–35. doi: 10.1016/j.pharmthera.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pignataro L, Miller AN, Ma L, et al. Alcohol regulates gene expression in neurons via activation of heat shock factor 1. J Neurosci. 2007;27(47):12957–66. doi: 10.1523/JNEUROSCI.4142-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varodayan FP, Pignataro L, Harrison NL. Alcohol induces synaptotagmin 1 expression in neurons via activation of heat shock factor 1. Neuroscience. 2011;193:63–71. doi: 10.1016/j.neuroscience.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci Res. 2004;49(4):379–85. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 76.Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12(12):893–9. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- 77.Bice PJ, Foroud T, Carr LG, et al. Identification of QTLs influencing alcohol preference in the High Alcohol Preferring (HAP) and Low Alcohol Preferring (LAP) mouse lines. Behav Genet. 2006;36(2):248–60. doi: 10.1007/s10519-005-9019-6. [DOI] [PubMed] [Google Scholar]
- 78.Newlin DB, Thomson JB. Alcohol challenge with sons of alcoholics: A critical review and analysis. Psychol Bull. 1990;108(3):383–402. doi: 10.1037/0033-2909.108.3.383. [DOI] [PubMed] [Google Scholar]
- 79.Kirstein SL, Davidson KL, Ehringer MA, Sikela JM, Erwin VG, Tabakoff B. Quantitative trait loci affecting initial sensitivity and acute functional tolerance to ethanol-induced ataxia and brain cAMP signaling in BXD recombinant inbred mice. J Pharmacol Exp Ther. 2002;302(3):1238–45. doi: 10.1124/jpet.302.3.1238. [DOI] [PubMed] [Google Scholar]
- 80.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 81.Moonat S, Starkman BG, Sakharkar A, Pandey SC. Neuroscience of alcoholism: molecular and cellular mechanisms. Cell Mol Life Sci. 2010;67(1):73–88. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;32(9):1525–34. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 83.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hua Z, Lv Q, Ye W, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sathyan P, Golden HB, Miranda RC. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: Evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J Neurosci. 2007;27(32):8546–57. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pietrzykowski AZ, Friesen RM, Martin GE, et al. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron. 2008;59(2):274–87. doi: 10.1016/j.neuron.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD. Up-regulation of MicroRNAs in brain of human alcoholics. Alcohol Clin Exp Res. 2011;35(11):1928–37. doi: 10.1111/j.1530-0277.2011.01544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mayfield RD. Human alcoholic brain: What have we learned and where are we going?; Annual Conference of the Australian Neuroscience Society; Gold Coast, Australia. 2012. [Google Scholar]
- 90.Crabbe JC. Review. Neurogenetic studies of alcohol addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363(1507):3201–11. doi: 10.1098/rstb.2008.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11(3-4):195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- 92.Baek MN, Jung KH, Halder D, et al. Artificial microRNA-based neurokinin-1 receptor gene silencing reduces alcohol consumption in mice. Neurosci Lett. 2010;475(3):124–8. doi: 10.1016/j.neulet.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 93.Jeanblanc J, He DY, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. J Neurosci. 2009;29(43):13494–502. doi: 10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lasek AW, Janak PH, He L, Whistler JL, Heberlein U. Downregulation of mu opioid receptor by RNA interference in the ventral tegmental area reduces ethanol consumption in mice. Genes Brain Behav. 2007;6(8):728–35. doi: 10.1111/j.1601-183X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 95.Lesscher HM, Wallace MJ, Zeng L, et al. Amygdala protein kinase C epsilon controls alcohol consumption. Genes Brain Behav. 2009;8(5):493–9. doi: 10.1111/j.1601-183X.2009.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rewal M, Jurd R, Gill TM, He DY, Ron D, Janak PH. Alpha4-containing GABAA receptors in the nucleus accumbens mediate moderate intake of alcohol. J Neurosci. 2009;29(2):543–9. doi: 10.1523/JNEUROSCI.3199-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thorsell A, Repunte-Canonigo V, O’Dell LE, et al. Viral vector-induced amygdala NPY overexpression reverses increased alcohol intake caused by repeated deprivations in Wistar rats. Brain. 2007;130(Pt 5):1330–7. doi: 10.1093/brain/awm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang AR, Liu J, Yi HS, et al. Binge Drinking: In Search of its Molecular Target via the GABA(A) Receptor. Front Neurosci. 2011;5:123. doi: 10.3389/fnins.2011.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hoplight BJ, Sandygren NA, Neumaier JF. Increased expression of 5-HT1B receptors in rat nucleus accumbens via virally mediated gene transfer increases voluntary alcohol consumption. Alcohol. 2006;38(2):73–9. doi: 10.1016/j.alcohol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 100.Worst TJ, Tan JC, Robertson DJ, et al. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res. 2005;80(4):529–38. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- 101.Carr LG, Kimpel MW, Liang T, et al. Identification of candidate genes for alcohol preference by expression profiling of congenic rat strains. Alcohol Clin Exp Res. 2007;31(7):1089–98. doi: 10.1111/j.1530-0277.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu Y, Ehringer M, Yang F, Sikela JM. Comparison of global brain gene expression profiles between inbred long-sleep and inbred short-sleep mice by high-density gene array hybridization. Alcohol Clin Exp Res. 2001;25(6):810–8. [PubMed] [Google Scholar]
- 103.Letwin NE, Kafkafi N, Benjamini Y, et al. Combined application of behavior genetics and microarray analysis to identify regional expression themes and gene-behavior associations. J Neurosci. 2006;26(20):5277–87. doi: 10.1523/JNEUROSCI.4602-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rulten SL, Ripley TL, Hunt CL, Stephens DN, Mayne LV. Sp1 and NFkappaB pathways are regulated in brain in response to acute and chronic ethanol. Genes Brain Behav. 2006;5(3):257–73. doi: 10.1111/j.1601-183X.2005.00157.x. [DOI] [PubMed] [Google Scholar]


