Fig. (2).
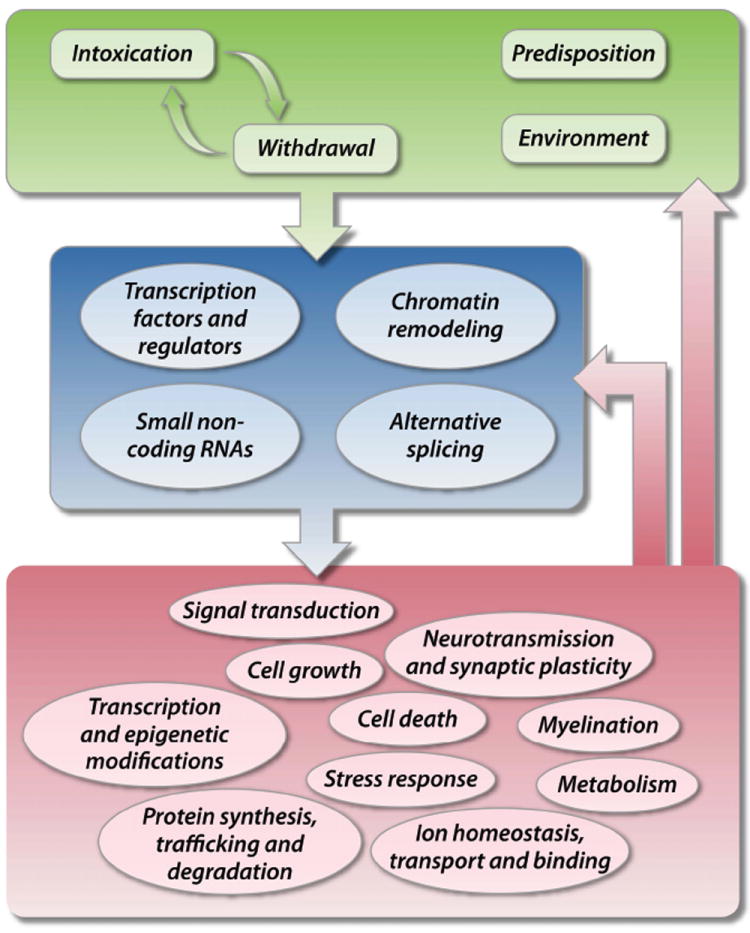
Schematic summary of molecular mechanisms hypothesized to drive the genome-wide regulation of gene expression in alcoholism. The green halo regroups potential causative factors for transcriptional changes in relation to alcoholism. Single nucleotide polymorphisms may influence the expression level of a given gene and the activity of its protein product, and thereby predispose an individual to increased vulnerability to or protection against alcoholism [2]. On the other hand, environmental variables, such as stressful life events or comorbid diseases, may also impact gene expression and thereby interact with the effects of ethanol [2]. The blue halo identifies molecular mechanisms that could mediate the global co-regulation of gene expression across the genome. Recent microarray and RNA-seq studies have pinpointed a potential role for transcription factors [37-40], epigenetic modifications [29, 35, 41, 42] and miRNAs [33, 34, 43] in the coordinated regulation of gene sets. In addition, RNA-seq is starting to uncover that alcoholism can affect the relative expression of different splice variants of a given gene [89]. The red halo highlights the functional categories that are enriched among genes differentially expressed in the brain of rodent predisposition models, rodent models of ethanol exposure and human alcoholics (see Table 2 for details). Changes in the expression level of genes encoding proteins involved in mRNA and small non-coding RNA processing, DNA methylation, histone acetylation and methylation can in turn contribute to accentuate, dampen or reorient the effects of the mechanisms identified in the blue halo. Moreover, changes in brain structure, connectivity and function resulting from the altered expression of genes involved in myelination, neurotransmission, signal transduction etc. are hypothesized to ultimately produce the behavioral impairments characteristic of alcoholism.
