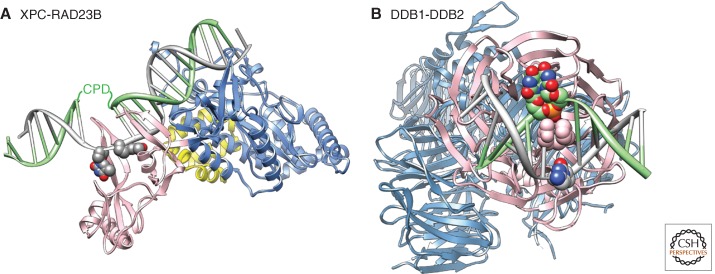
Figure 2.
Structural basis for damage recognition in NER. (A) Structure of RAD4(XPC)-RAD23 bound to a CPD lesion. The TDG/BHD1 domains (blue) of RAD4/XPC bind to undamaged DNA, the BHD2/BHD3 domains (pink) encircle two nucleotides in the nondamaged strand of DNA. DNA is represented in gray with the two thymidine residues opposite the lesion in atom color. The position of the CPD lesion that is disordered in this structure is indicated. A fragment of RAD23 is shown in yellow. Note that the CPD lesion is present in a two base pair mismatch in this structure. The damaged DNA strand is shown in green, the undamaged strand in gray. The figure was made using the Chimera extensible molecular modeling system located at UCSF (www.cgl.ucsf.edu/chimera), using the structure PDB 2QSG (Min and Pavletich 2007). (B) Structure of DDB1-DDB2 bound to a CPD lesion. The UV-DDB complex, made up of DDB1 (light blue) and DDB2 (pink), binds DNA (gray) containing a CPD (atom color, green) through the DDB2 subunit. The DNA-binding site in DDB2 is located on one site of the β-propeller; DDB1 binds DDB2 on the opposite side. A wedge made up of residues F371, Q372, and H373 in DDB2 (shown in pink van der Waals representation) inserts into the DNA helix at the lesion site from the minor groove. The two modified nucleotides of the 6-4PP insert into a shallow binding pocket in DDB2. The damaged DNA strand is shown in green, the undamaged strand in gray. The figure was made using the Chimera extensible molecular modeling system located at UCSF (www.cgl.ucsf.edu/chimera), using the structure PDB A408 (Fischer et al. 2011).