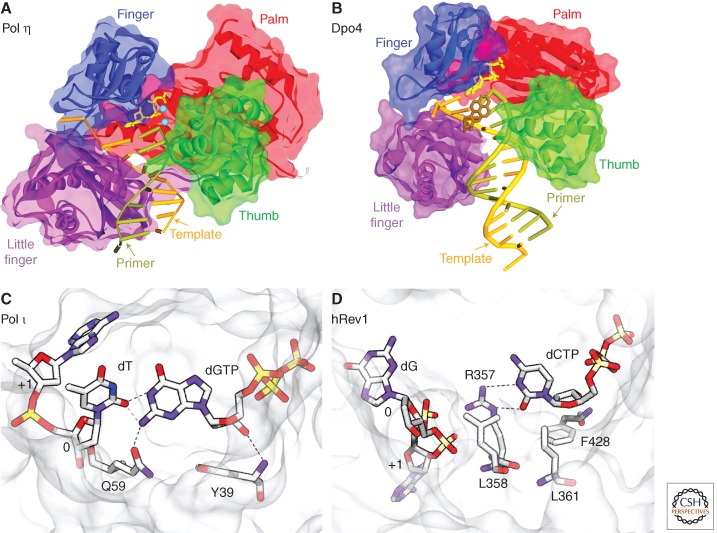
Figure 3.
Structural insights into TLS polymerases and their mutagenic specificity. In panels A and B, the main domains are color-coded: (red) palm; (green) thumb; (blue) fingers; (purple) little finger. (A) Crystal structure of human Pol η in a ternary complex with a CPD. In this view, the 3′T of the CPD is in the active site and is correctly paired with incoming dATP (PDB: 3MR3) (Biertümpfel et al. 2010). (Rust) The template strand; (olive green) the primer; (yellow) the incoming dNTP. (Burgundy stick) The position of the CPD; (small blue spheres) the metal ions. The protein backbone is represented by the ribbon surrounded by the semitransparent solvent-accessible surface. (B) Crystal structure of the S. solfataricus Dpo4 in a ternary complex with DNA containing a benzo[a]pyrene lesion (brown) and incoming nucleotide (yellow) (PDB: 1S0M_BP-2) (Ling et al. 2004b). As can be seen, the benzo[a]pyrene lesion is flipped into the major groove, so as to accommodate base pairing. (C) Human Pol ι making a G:T mispair (PDB:3GV8); note that the template dT and incoming dG are both in an anti conformation (Kirouac and Ling 2009) and the mispair is stabilized through hydrogen bonds with Gln59. (D) Arg357-directed dC incorporation by human Rev1 (PDB: 3GQC) (Swan et al. 2009). (C,D) (White/gray) The protein surface; (dotted lines) hydrogen bonds.