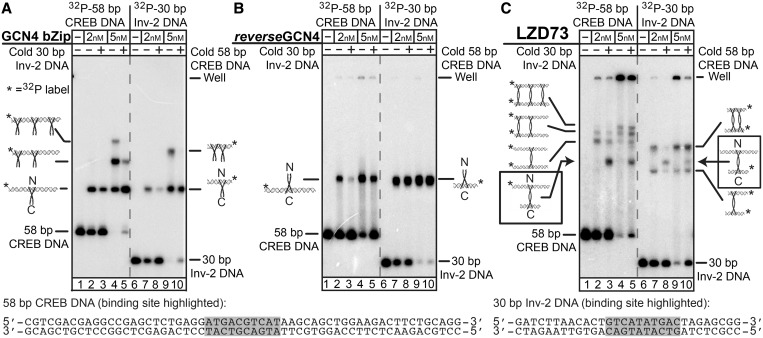
Figure 2.
Demonstration of LZD73:DNA sandwich complexes by EMSA. In each panel, lanes 1–5 contain 1 nM 32P-labeled 58-bp CREB DNA, with 1 nM unlabeled 30-bp Inv-2 DNA in lanes 3 and 5. Lanes 6–10 have 1 nM 32P-labeled 30 bp Inv-2 DNA, with 1 nM unlabeled 58-bp CREB DNA in lanes 8 and 10. Cartoons show binding ratios do not imply details of non-specific protein:DNA structures. (A) EMSA with the GCN4 bZip single-binding control peptide shows a weak preference for CREB DNA, and non-specific protein binding is seen when protein is in excess. (B) EMSA with the single-binding reverseGCN4 peptide, with a C-terminal DNA-binding domain shows a strong preference for binding to Inv-2 over CREB and a much lower extent of non-specific binding than GCN4. (C) EMSA with LZD73 shows complexes with lower mobility, suggesting they are sandwich complexes, and the presence of both DNAs in the same complex in the bands indicated with the arrows confirms binding of two DNAs to one peptide. Identification of 58/58 and 30/30 sandwiches is by comparative mobility. Bands that appear only with increased protein concentration are assigned to non-specific binding.