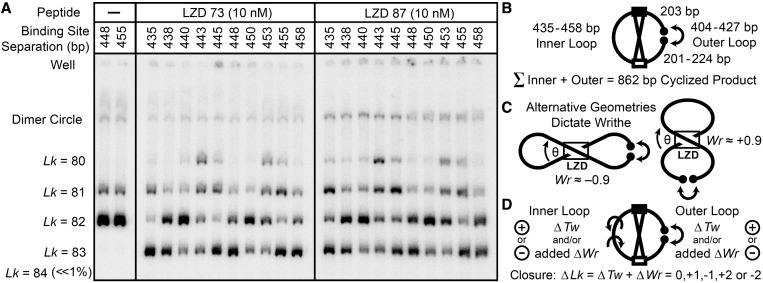
Figure 4.
Topological effects of protein-induced looping, for ten 862 bp cyclized DNA fragments with CREB and Inv-2 sites separated by 435–458 bp. DNA (0.3 nM) was cyclized by T4 DNA ligase in the presence of LZD73 or LZD87, treated with BAL31 nuclease and analyzed as in Figure 3. Cyclization of DNA alone is shown for the 448 and 455 constructs on the left; all the other constructs give the same results. The distribution of topoisomers in the presence of LZD peptides, including new positive and negative supercoils, is a periodic function of binding site separation. (B) Schematic of the variation of inner and outer loop lengths at a constant 862 bp total length. (C) Looping introduces a writhe crossover, which can be of either sign because the binding sites are palindromic. The peptide can also introduce a local ΔTw. (D) A ΔTw in the inner lobe is required to bring binding sites into alignment to form a protein-mediated loop, and a ΔTw in the outer loop is required for ring closure. Both twist changes and any writhe in the lobes contribute to the total observed ΔLk.