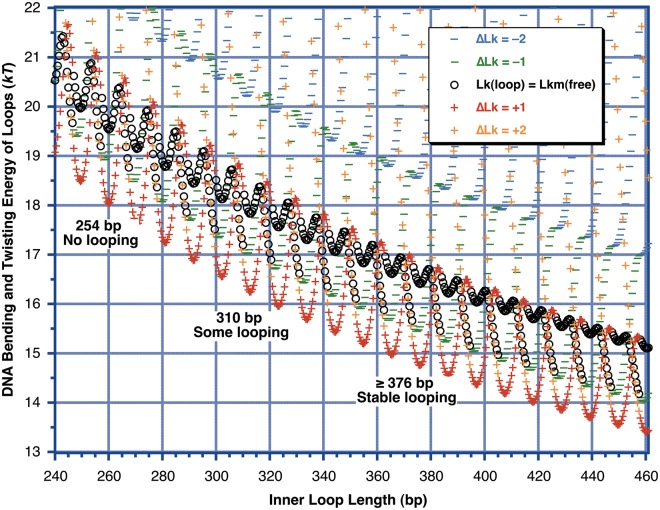
Figure 7.
Calculated DNA strain energy for different loop-containing minicircle topoisomers as a function of loop length (inner loop binding site separation), using the model of Figure 5 and the worm-like coil as in Figure 6. The ΔLk is defined relative to the Lkm of free DNA at each DNA length. The outer loop length is held fixed at 414 bp as in the experiment of Figure 3, and the qualitative results of Figure 3 are noted within the figure. All model parameters were those for LZD87 from Table 1 except that no offset was included. The energies of all of the topoisomers seem to be discontinuous because the reference Lkm of the free DNA changes by 1 abruptly as the DNA length crosses (n + ½) helical turns. Because of the large writhe induced on looping, the topoisomer with Lk equal to the Lkm of free DNA never corresponds to the lowest-energy topoisomer. In this model, the overall minimum possible energy, where there is no twist strain and the total strain energy is equal to the bending energy, occurs for helically out-of-phase sites. This initially surprising result arises because for the protein geometry assumed here the out-of-phase sites can be brought together with no change in twist for the inner loop. This introduces a change of ΔTw = (−ΔWr) in the outer lobe, which will then relax by the introduction of a twist change of ΔTw = +ΔWr ≈ ±1 to return the outer lobe to its relaxed twist. The end result is a loop with almost no twist strain, with ΔLk = ±1 ≈ +ΔWr relative to relaxed DNA. ΔLk = +1 is lower in energy than ΔLk = −1 because the best-fit value θ = 33° leads to a lower bend energy for the Wr = +0.9 geometry.