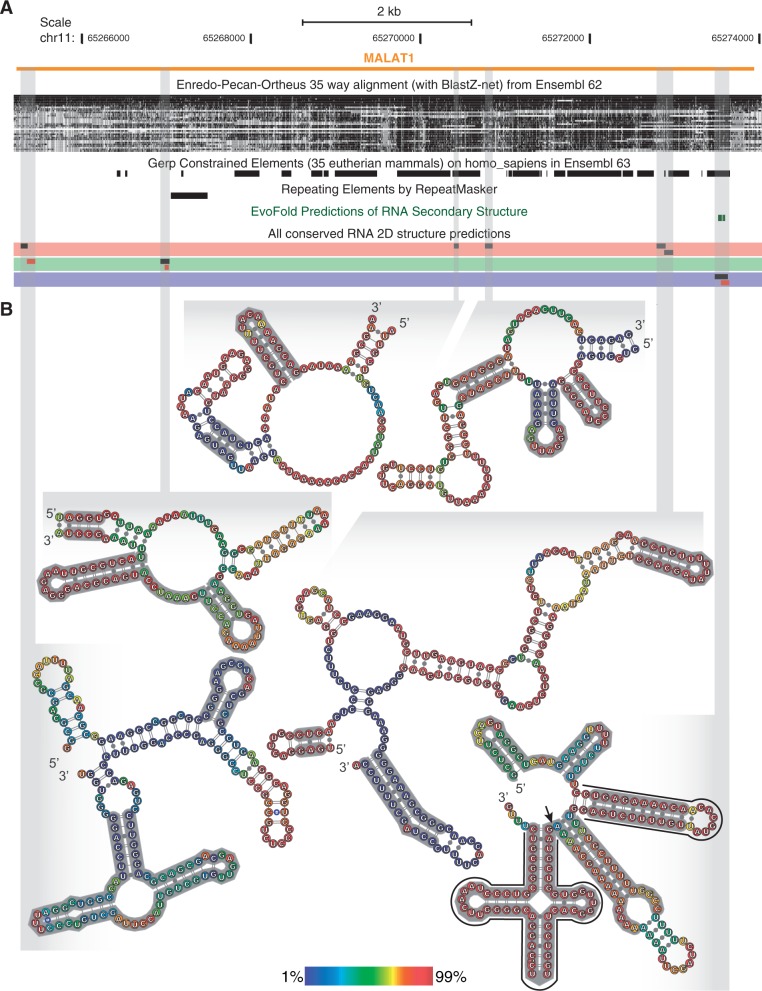
Figure 5.
Structural characterization of the long ncRNA MALAT1. (A) UCSC genome browser (hg19) screenshot of the MALAT1 locus with the following tracks: EPO multiple genome alignment (used to emit predictions), GERP++ constrained sequence element track, repeat elements, EvoFold evolutionarily conserved RNA secondary structure predictions, and the ECS predictions reported herein, with colors representing the algorithm used to make the prediction (SISSIz in red, SISSIz with RIBOSUM in green, RNAz in blue). Red rectangles represent ECS predictions that are structurally congruent with the reference sequence. (B) Human RNA secondary structures associated to predictions. Consensus RNA secondary structures were extracted from the associated alignments and used as a constraint for folding the human sequence with RNAfold (76). The base colors represent the (unconstrained) partition function base-pairing probabilities associated to the represented structures. Gray structure annotations correspond to RNAalifold consensus structures supported by conserved or compensatory mutations. The substructures outlined in black (bottom right) correspond to the previously characterized mascRNA and associated stem-loop, which are required for efficient RNAse P cleavage (cleavage site indicated with an arrow) (74). Structure representations were created with VARNA (77).