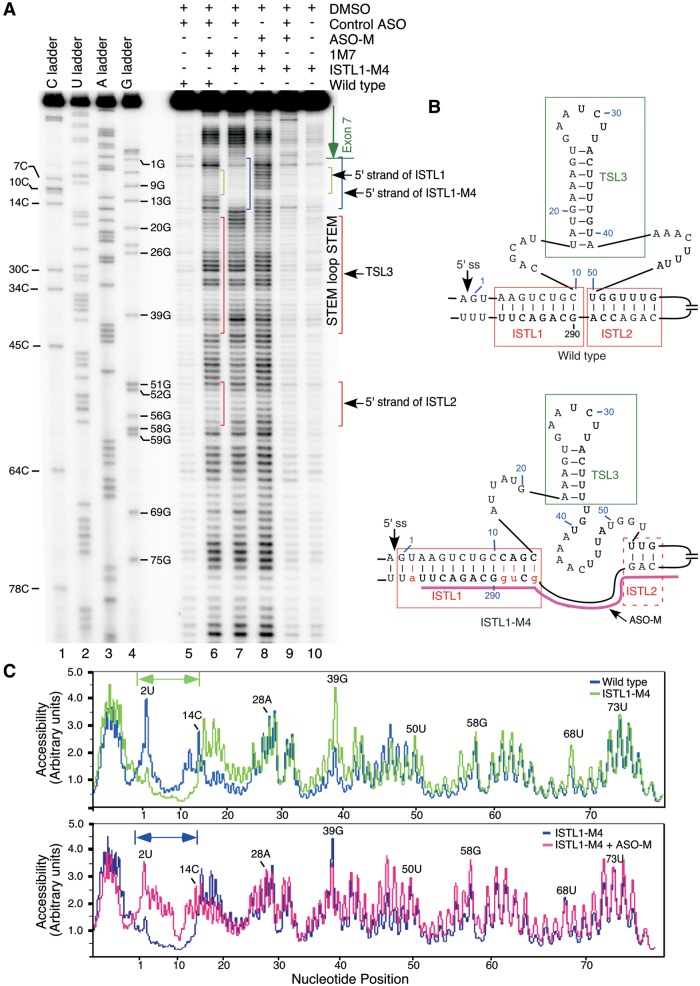
Figure 7.
Validation of the engagement of the 5′ strand of ISTL1 in structure formation. (A) SHAPE results for the 5′ portion of intron 7 of the wild-type and ISTL1-M4 mutant RNAs generated using Primer#10. Based on the sequencing ladders, positions of residues and locations of structures are marked on the gel. (B) Abridged mfold predicted structure of SMN2 intron 7. The probed structure is in agreement with the mfold predicted structure. Nucleotide substitutions are shown in small-case letters and highlighted in red. Annealing site of ASO-M is indicated. Abbreviations are the same as in Figure 6. (C) Alignment of raw peak profiles for the wild-type and mutant RNAs. Nucleotides that constitute the 5′ strand of ISTL1 in the mutant RNA are marked. Peak profiles were generated using MultiGauge Software version 3.0 (FujiFilm).