Figure 1.
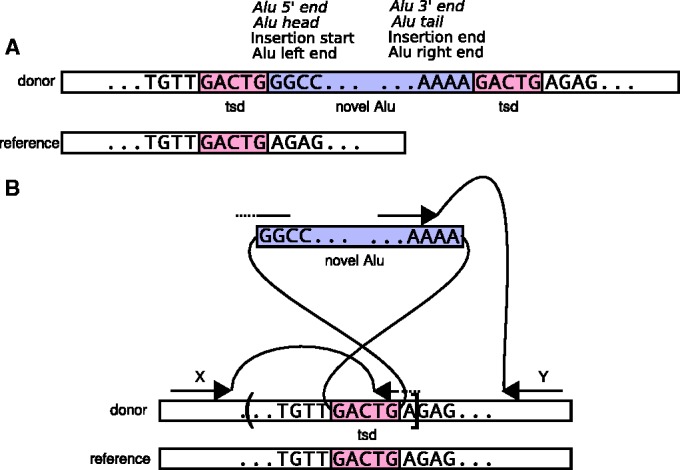
Detecting Alu insertions and TSDs. (A) Genome after an Alu insertion on the positive strand. The human reference genome is at the bottom. The newly sequenced ‘donor’ genome with an Alu insertion and a TSD is above. Above is the terminology. ‘Head’ and ‘tail’ refer to the inner sequence of the Alu, whereas ‘left’, ‘right’, ‘insertion start’ and ‘insertion end’ refer to the orientation of the genome. If the Alu were on the negative strand, the terms in italic would be flipped. (B) Detection of Alu insertions. The donor genome is shown aligned to the reference genome. Because of the TSD, the left end of an Alu will start at the right end of the TSD. Two read pairs supporting the insertion are shown. Read pair X has a split-mapped read aligned across the 5′ (left) end of the Alu (GGCC), and the right end of the TSD, directly identifying the breakpoint. Read pair Y supports the presence of an insertion, but does not identify the exact breakpoint (other end of the TSD). As only the left endpoint is detected (between G/A), the right end of the confidence interval is the A following the breakpoint, whereas the left is only estimated. The detected breakpoint is represented by a square bracket, and the undetected one by a round bracket.
