Table 3.
WGS versus Exome Capture
| Region | GS | XS | 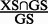 |
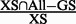 |
|---|---|---|---|---|
| Coding exons | 12 | 10 | 5/12 = 0.42 | 8/10 = 0.80 |
| Exons | 25 | 11 | 6/25 = 0.24 | 9/11 = 0.82 |
| Probes | 12 | 12 | 9/12 = 0.75 | 11/12 = 0.92 |
Coding exons, coverage  10× 10× |
9 | 10 | 5/9 = 0.56 | 8/10 = 0.80 |
Exons, coverage  10× 10× |
11 | 11 | 6/11 = 0.55 | 9/11 = 0.82 |
Probes, coverage  10× 10× |
12 | 12 | 9/12 = 0.75 | 11/12 = 0.92 |
GS: calls from WGS HTS data, with CI 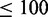 .
.
XS: calls from exome capture HTS data, with CI 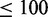 . All-GS: calls from WGS data, without the CI filter. These are proximal to the genomic features but may not directly intersect them. XS
. All-GS: calls from WGS data, without the CI filter. These are proximal to the genomic features but may not directly intersect them. XS  GS/GS: calls made with WGS data in the corresponding region that were also found with exome capture data in that region, i.e. ‘recall’ of exome capture relative to WGS. XS
GS/GS: calls made with WGS data in the corresponding region that were also found with exome capture data in that region, i.e. ‘recall’ of exome capture relative to WGS. XS  All-GS/XS: calls made with exome capture data in that region that match some call made with WGS data, i.e. ‘precision’ of exome capture relative to WGS. For Exons and Coding Sequence, we include the full set of regions from our annotation, regardless of whether they are captured by the kit. Bottom rows: Genomic calls are restricted to regions where exome capture data have coverage of at least 10×. Coverage was computed by tiling the genome with 100 bp windows. All regions are extended by 50 bp.
All-GS/XS: calls made with exome capture data in that region that match some call made with WGS data, i.e. ‘precision’ of exome capture relative to WGS. For Exons and Coding Sequence, we include the full set of regions from our annotation, regardless of whether they are captured by the kit. Bottom rows: Genomic calls are restricted to regions where exome capture data have coverage of at least 10×. Coverage was computed by tiling the genome with 100 bp windows. All regions are extended by 50 bp.
