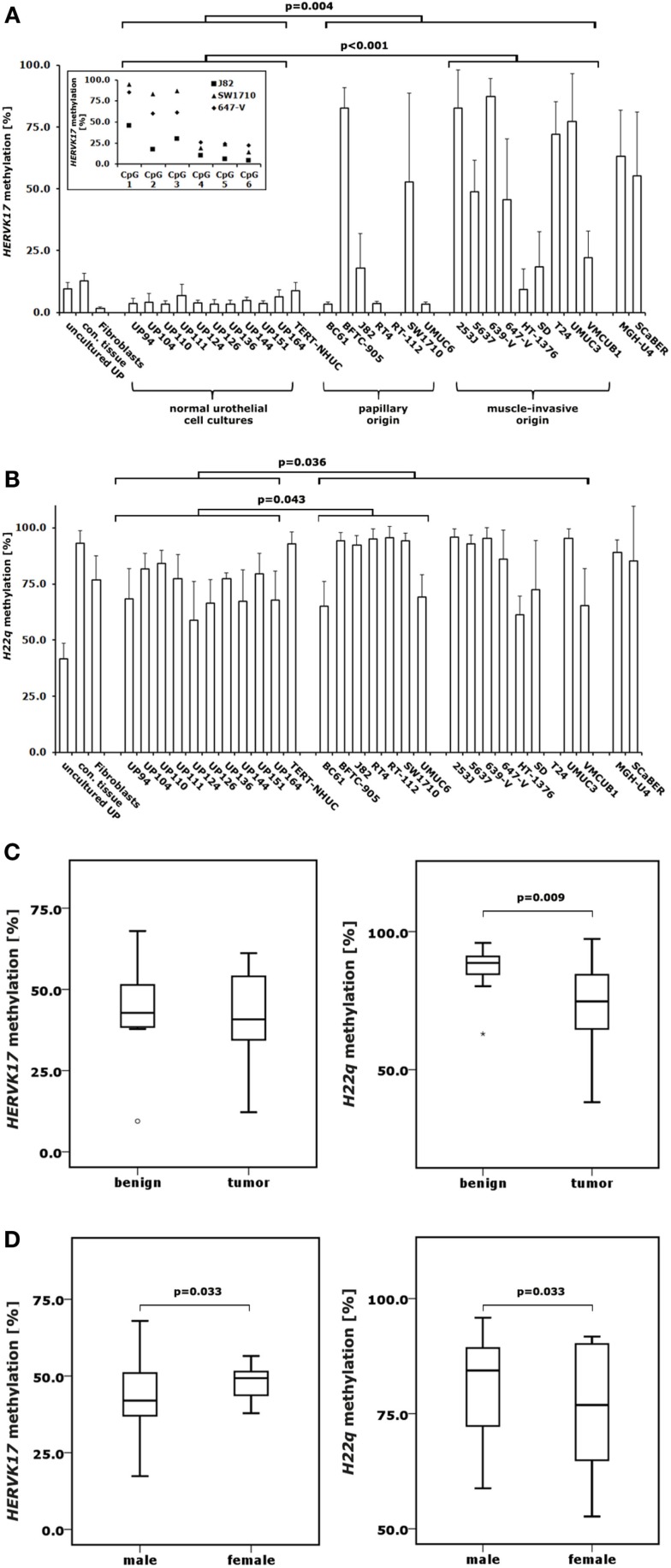
Figure 3.
DNA methylation changes in proviral HERVK17 and H22q LTRs in bladder cancer. DNA methylation in the LTRs of HERVK17 (A) and H22q (B) were analyzed by pyrosequencing in 10 normal urothelial cell cultures and 18 bladder cancer cell lines. Additionally, HERVK17 and H22q DNA methylation was assessed in immortalized urothelial cells (TERT-NHUC) and in uncultured epithelial cells (uncultured UP) and connective tissue from one ureter. (C) DNA methylation of HERVK17 and H22q LTRs were each analyzed in a set of 12 benign and 23 cancerous bladder tissues. (D) DNA methylation of HERVK17 and H22q LTRs from tumor samples were each plotted against patients’ gender. Methylation is plotted as mean methylation value from six CpGs each in percent. The high standard deviation in some samples results from differences in the methylation within the HERVK17 sequence, where the first three CpGs are generally higher methylated as exemplified for data from J82, SW1710, and 647-V bladder cancer cell lines in the insert (A). p Values calculated by the Mann–Whitney U-test are given above the brackets for significant changes (p < 0.05). Missing p values demonstrate changes without reaching the level of significance.