Abstract
Deubiquitinases (DUBs) play an important role in regulating the ubiquitin landscape of proteins. The DUB AMSH (Associated Molecule with the SH3 domain of STAM) has been shown to be involved in regulating the ubiquitin-dependent downregulation of activated cell surface receptors via the endo-lysosomal degradative pathway. Therefore small molecule AMSH inhibitors will be useful chemical probes to study the effect of AMSH DUB activity on cell surface receptor degradation. Currently there are no known selective inhibitors of AMSH or high-throughput compatible assays for their identification. We report the development and optimization of a novel FRET based add-and-read AMSH deubiquitinase assay in a 384-well format. In this format, the optimal temperature for a high throughput screen (HTS) was determined to be 30°C, the assay tolerates 5% DMSO and has a Z-score of 0.71 indicating HTS compatibility. The assay was used to show that AMSH selectively cleaves Lys63-linked diubiquitin over Lys48- and Lys11-linked diubiquitin. The IC50 value of the non-specific small molecule DUB inhibitor N-ethylmaleimide was 16.2 ± 3.2 μM and can be used as a qualitative positive control for the screen. We conclude that this assay is HTS compatible and can be used to identify novel small molecule inhibitors of AMSH.
Keywords: HTS, FRET, deubiquitinase assay, AMSH
Introduction
Ubiquitination of proteins has been implicated in numerous biological pathways including, but not limited to, cell cycle regulation, DNA damage, and endocytosis [1–4]. Ubiquitin molecules (Ub) are ligated to their target proteins as both mono- and polyubiquitin chains. The diversity in the linkages (eight different linkages) in the polyubiquitin chains facilitates the relay of a variety of signals [4,5]. Deubiquitinases (DUBs) cleave the isopeptide bond in the polyubiquitin chain or the protein-Ub linkage to further regulate Ub mediated signaling [4,6]. DUBs are part of multi-protein complexes and have both enzymatic and scaffolding functions. Their knockdown not only eliminates the enzymatic function but also disrupts the scaffolding functions resulting in the dysfunction of the entire complex. DUB activity specific small molecule inhibitors will provide the precision to specifically study the role of enzymatic functions within these multi-protein complexes. Recent studies have also implicated DUBs in several diseases, particularly cancer, and targeting DUBs for therapeutic intervention is an emerging theme [7,8].
Associated molecule with the SH3 domain of STAM (AMSH) plays a key role in regulating receptor sorting at the endosome through its function as a deubiquitinase [9–11]. AMSH belongs to the JAMM (JAB1/MPN/MOV34) deubiquitinase family and specifically cleaves Lys63-linked polyubiquitin [12,13]. Through interactions at multiple points in endocytic cargo sorting, AMSH plays a critical regulatory role in cell surface receptor downregulation [14,15]. Downregulation is accomplished through the recognition of specific ubiquitination patterns on internalized receptors, specifically multi-monoubiquitination and Lys63 polyubiquitination [3, 11]. Spatial and temporal dysregulation of AMSH-mediated deubiquitination of internalized ubiquitinated cell surface receptors affects their sorting to the lysosome. Consistently, knockdown of endogenous AMSH or overexpression of catalytically inactive AMSH mutants has been shown to promote the lysosomal degradation of epidermal growth factor receptor (EGFR) as well as other cell surface receptors [16–22]. Small molecule inhibitors of AMSH will be valuable chemical probes for dissecting endocytic cargo sorting. Currently there are no known inhibitors of AMSH and no report of a high-throughput compatible assay for the identification of potential inhibitors.
AMSH alone has been shown to have deubiquitinase activity in cell-free assays, making it suitable for high-throughput screens [13,23–25]. There are many assay types used in high-throughput formats to identify inhibitors of enzymes, such as proteases [26,27]. The ease of execution and cost-per-well has made fluorescence based assays a popular choice in the HTS community. Our lab previously reported the development of fluorescence polarization assays and high-throughput screens to identify inhibitors of protein-protein interactions [28–30]. Since it is known that AMSH cleaves Lys63 ubiquitin chains, we chose to explore a fluorescence resonance energy transfer (FRET) based system [13,23–25]. In a typical FRET assay, the donor and acceptor/quencher are spaced by a suitable linker which when cleaved results in the loss of FRET/gain in fluorescence. The catalytic domain of AMSH and a Lys63-linked diubiquitin probe labeled with a donor and a quencher on different ubiquitins was used in this study. The development and optimization of a FRET based high-throughput compatible AMSH assay is reported. Importantly, this assay can be easily modified for other deubiquitinases that demonstrate linkage specific cleavage, which may not readily cleave other commercially available ubiquitin probes.
Materials and Methods
General Reagents
All FRET labeled diubiquitin probes were purchased from BostonBioChem (R&D) and were stored in 50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM DTT. The catalytic domain of AMSH (residues 219 to 424) was expressed and purified as previously described and stored in 50 mM Tris pH 7.4, 50 mM NaCl, 1mM DTT [24]. All concentrations shown in parenthesis are final assay concentrations. Anti-Ubiquitin antibody (P4D1) was obtained from Cell Signaling.
Deubiquitinase assay
All measurements were made on 384-well, low-volume, black round-bottom polystyrene NBS microplate (Corning) using a Spectramax M5 plate reader (MDS). All reactions were done in reaction buffer, 50 mM HEPES pH 7.0, 25 mM KCl, 5 mM MgCl2, 1 mM DTT, in a final volume of 20 μL [24]. Measurements were taken at an excitation wavelength of 544 nm and an emission wavelength of 572 nm and cutoff was set at 570 nm. The reported fluorescence values (RFU) are a ratio of total fluorescent signal to the background obtained from the probe alone. All experiments were performed at least twice and each time as duplicates.
Dose-response and time-dependent studies of AMSH and K63POS1 probe
AMSH was titrated (15.6 nM – 250 nM) into a constant concentration of K63POS1 probe (500 nM) and the deubiquitinase reaction was monitored over time at 15 min intervals for 2 h at 30°C. Optimal probe concentration was determined by titrating K63POS1 probe (200 nM – 600 nM) while holding the concentration of AMSH constant at 125 nM. The plate was read following a 90 min incubation. The relationship between product formation and fluorescence was determined by incubating 10 μM of K63POS1 probe with 5 μM AMSH overnight at room temperature. Cleaved probe was then diluted 2-fold in reaction buffer followed by fluorescent measurements.
Dimethyl sulfoxide (DMSO) tolerance
Increasing concentrations of dimethyl sulfoxide (0.5 – 25% of the assay volume 20 μL) was added to AMSH (125 nM) and the mixture was incubated for 30 min. K63POS1 probe (500 nM) was then added to the reaction mixture and incubated for an additional 90 min at 30°C and measurements were made. Statistics were performed using a Student t-test.
NEM inhibition assay
Increasing concentrations of NEM (0.8 μM – 500 μM) were added to AMSH (125 nM) with a 1:9 ratio of volumes and incubated at room temperature for 30 min. 10 μL of K63POS1 probe (500 nM) was then added to bring the assay volume to 20 μL. Fluorescence measurements were made after a 90 min incubation at 30°C. The data was fitted and the IC50 values were derived using non-linear least square fit to a single site-binding model (SigmaPlot 11.0). NEM time-dependent inhibition was performed by incubating AMSH (125 nM) with 15 μM or 30 μM NEM for 1 h, 45 min, 30 min or 15 min at room temperature. The K63POS1 probe was then added an incubated for 90 min at 30°C.
The affect of NEM on the K63POS1 probe fluorescence was determined by incubating cleaved or uncleaved probe with NEM (0 – 500 μM) for 90 min at 30°C. Cleaved probe for this study was generated as described in dose-response and time-dependent studies of AMSH and K63POS1 probe section.
Z-score
AMSH (125 nM) was incubated for 30 min with or without 25 μM NEM at room temperature. K63POS1 (500 nM) probe was then added to AMSH, AMSH + NEM, and buffer only wells for 90 min at 30°C before taking fluorescence measurements. Plates were then sealed and incubated at 4°C overnight for the 24 hr measurement. Data from the 384-well plates were collected on separate days with 48 data points for each condition per day. Z-score was calculated using AMSH + K63POS1 probe as the negative control and K63POS1 probe alone as the positive control.
Results and Discussion
Selective cleavage of Lys63-linked diubiquitin probe by AMSH
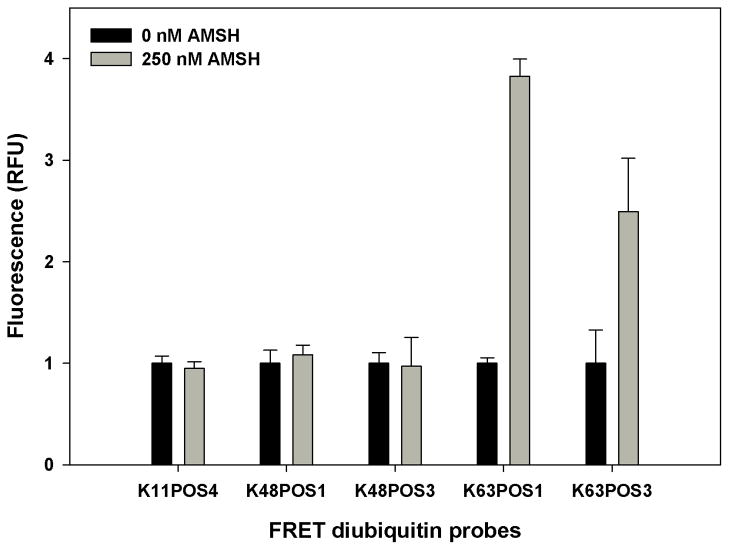
Several deubiquitinases have been shown to possess linkage specific cleavage, including members of the JAMM family [5]. AMSH has been shown to selectively cleave Lys63-linked polyubiquitin chains [13,23–25]. The catalytic domain of AMSH and a diubiquitin FRET probe in which the donor and quencher are placed on different ubiquitins linked by the Lys63 isopeptide bond was used to establish the AMSH assay. To identify a suitable probe and demonstrate AMSH’s cleavage selectivity, the catalytic domain of AMSH was incubated with five different diubiquitin probes (K11POS4, K48POS1, K48POS3, K63POS1 and K63POS3) that have three different linkages (Lys11, Lys48, and Lys63) (Fig. 1A–C). The figure shows the distance between the closest and farthest Lys residues on adjacent ubiquitins. Cleavage of the probes was assessed by the change in fluorescence following a 1 h incubation with AMSH (Fig. 2). The cleavage was also confirmed by SDS-PAGE (Fig. S1). No change was observed in the fluorescence reading in the presence of AMSH (Fig. 2, gray bars) with K11POS4, K48POS1 and K48POS3 probes compared to probe alone (Fig. 2, black bars) indicating AMSH did not cleave the Lys11- or Lys48- linked probes. A ~4- and ~3-fold increase in the fluorescence was observed with the K63POS1 and K63POS3 probes, respectively. This increase in fluorescence demonstrates that Lys63-linked diubiquitin is cleaved by AMSH. The difference in the fold-change between the two Lys63 probes is likely due to the different positions of the donor/quencher pair, wherein POS3 may interfere with cleavage by AMSH (Fig. 1C). K63POS1 probe had the larger signal window and therefore was used as the probe for all subsequent studies. The selectivity of substrate from the FRET study were consistent with the observed cleavage by Western blot analysis and previously reported studies that show AMSH preferentially cleaves Lys63-linked polyubiquitin chains over Lys48-linked polyubiquitin chains [13–15,23–25].
Figure 1.
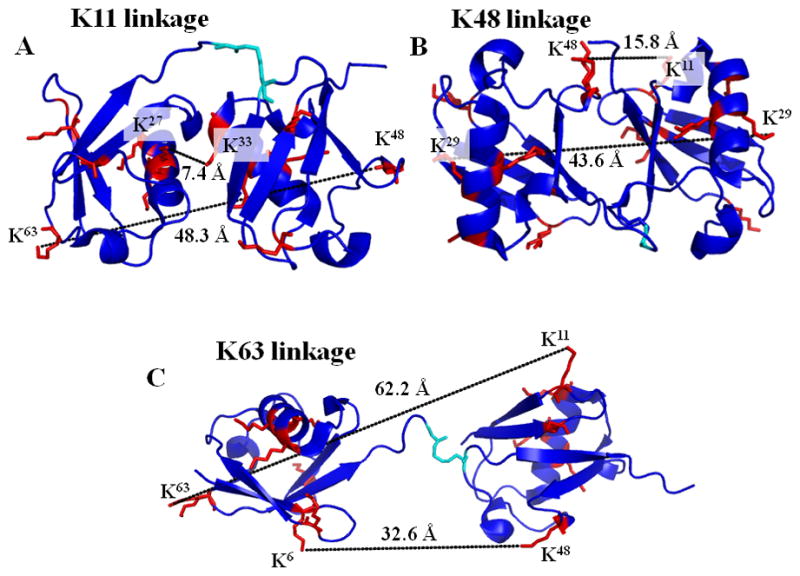
Diubiquitin structure. (A) Structure of K11 diubiquitin, (B) of K48 diubiquitin and (C) of K63 diubiquitin, with the lysine residues (red) and the isopeptide bond (cyan) shown as sticks. The largest and smallest distances between lysine residues on the proximal and distal ubiquitins are shown.
Figure 2.
AMSH probe selection. Diubiquitin probes of different linkages (final concentration 500 nM) were incubated with (gray bars) or without (black bars) 250 nM AMSH for 30 min at 30°C. (n = 2)
AMSH FRET assay optimization
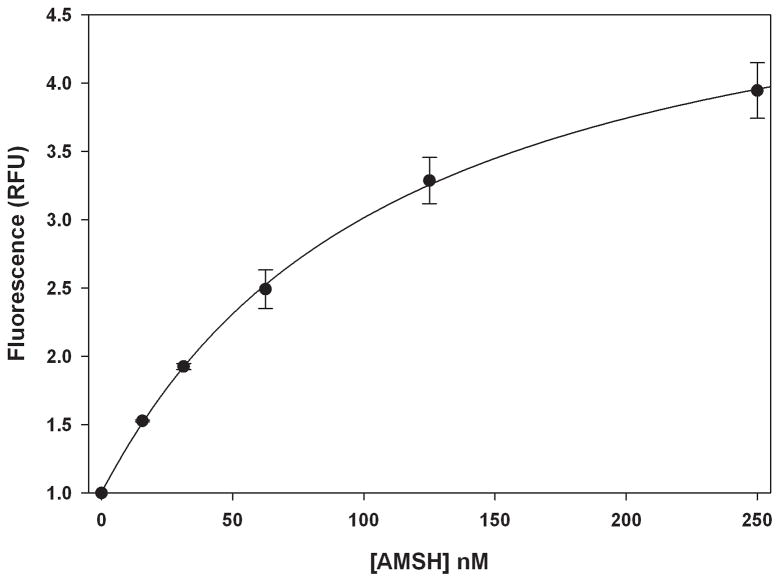
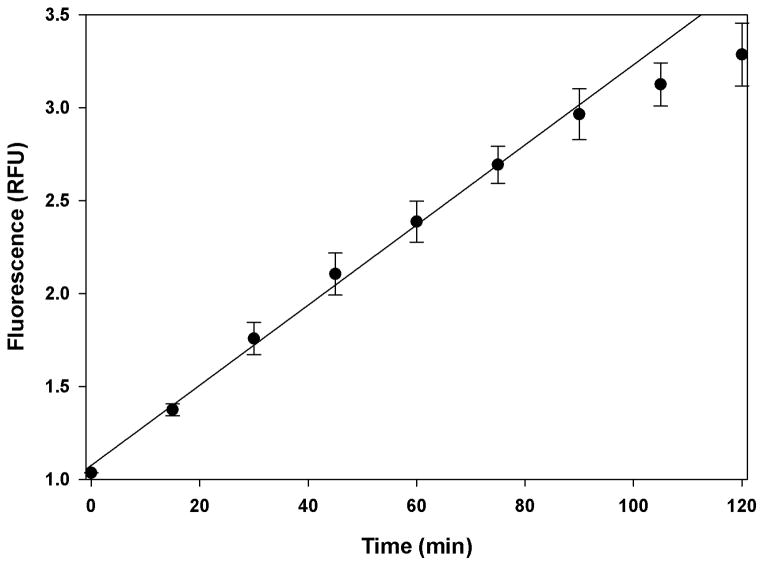
Next, to optimize the concentration of reagents and time of incubation to be used for a screen, we conducted dose-response and time-dependent studies. A dose-dependent increase in the fluorescence was observed with increasing concentrations of AMSH (0–250 nM) with the signal beginning to saturate at 250 nM (Fig. 3). For subsequent studies we selected 125 nM AMSH as this provided a reasonable signal window while minimizing reagents. The time-dependent increase in the cleavage of the K63POS1 probe was monitored over 2 h (Fig. 4). The fluorescent signal is linear up to 90 min after which it begins to plateau, due to complete consumption of K63POS1 probe. Therefore for the above concentrations of enzyme and substrate we concluded that an incubation time of 90 min will yield the largest signal window while still in the linear range. Next we performed a dose-dependent study with the K63POS1 probe. We observed increased signal with increasing K63POS1 probe concentration, which plateaued at 500 nM at 90 min (Fig. 5A, grey bars). To determine if this saturation was an artifact of the detection system, the K63POS1 probe was incubated with AMSH overnight to generate cleaved ubiquitin, which was then titrated in reaction buffer and fluorescence measured. We observed a linear relationship between probe concentration and fluorescence (Fig. 5B) suggesting that the saturation observed in Fig. 5A is not an artifact of the detection system. In this study, we also observed a smaller dose-dependent increase in the noise with increasing K63POS1 probe concentration (Fig. 5A, black bars). Although the best signal to noise (~4) was observed at both 200 and 500 nM of the K63POS1 probe, for subsequent studies we selected the 500 nM concentration as it provided a larger screening window (~9 vs. ~3).
Figure 3.
Fluorescence signal is dose-dependent. AMSH was titrated (0–125 nM) into 500 nM K63POS1 probe and incubated 2 h at 30°C. (n = 2)
Figure 4.
Fluorescence signal is time-dependent. 125 nM AMSH was incubated with 500 nM K63POS1 probe at 30°C. Signal was monitored over 2 h. (n = 2)
Figure 5.
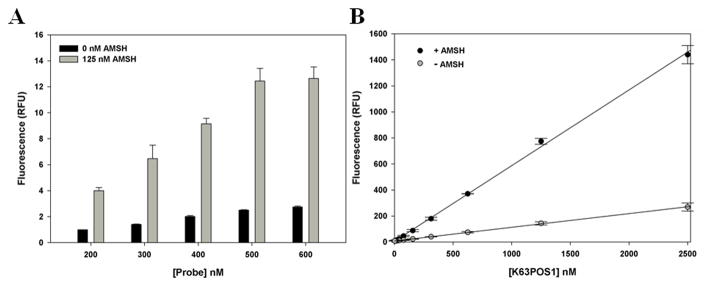
(A.) Optimization of K63POS1 probe concentration. AMSH concentration was held constant at 125 nM and K63POS1 probe was titrated in and incubated at 30°C for 90 min. (n = 2) (B.) Linear relationship between monoubiquitin formation and fluorescence. 10 μM of K63POS1 probe was incubated with or without 10 μM AMSH overnight. Cleaved probe was then titrated in buffer (2.5 nM – 2.5 μM). (n = 2)
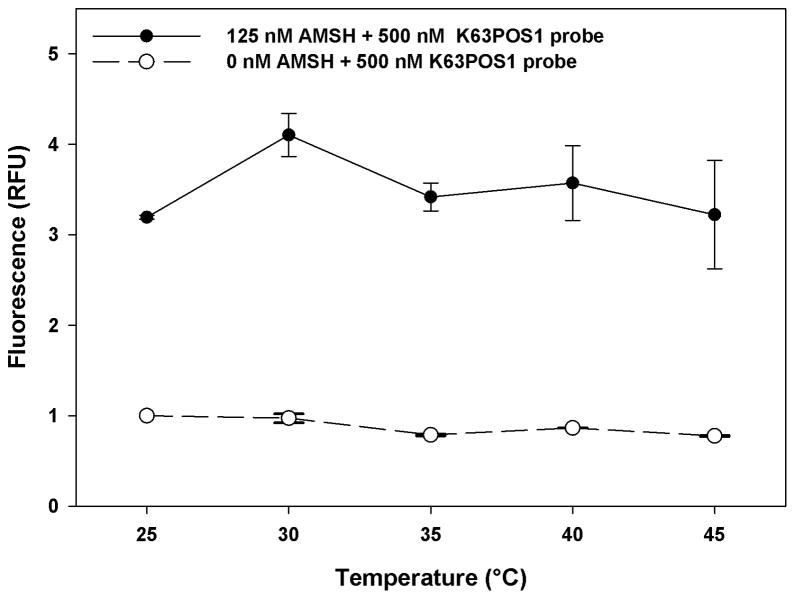
Temperature dependence
Screening large libraries takes several days to complete and despite the best climate controlled environment there are always fluctuations, therefore we next assessed the effect of temperature on the stability of this assay. This was accomplished by incubating the K63POS1 probe with and without AMSH at five different temperatures (25–45°C) for 90 min (Fig. 6). With increasing temperatures we observe a significant increase in the variability of the AMSH + K63POS1 fluorescent signal, particularly at temperatures above 40°C, but little change in probe alone. This indicates that temperatures should not exceed 40°C during a HTS. Additionally, we observe variation in the signal window with the largest window seen at 30°C. Although at 25°C, we observed less fluctuation in the signal with AMSH we conclude that 30°C is optimal as it results in a larger screening window in this assay format.
Figure 6.
Temperature dependence of AMSH deubiquitinase activity. K63POS1 probe (500 nM) was incubated either with or without AMSH (125 nM) for each temperature. For each temperature, measurements were taken following a 90 min incubation of AMSH and probe. (n = 2)
DMSO tolerance
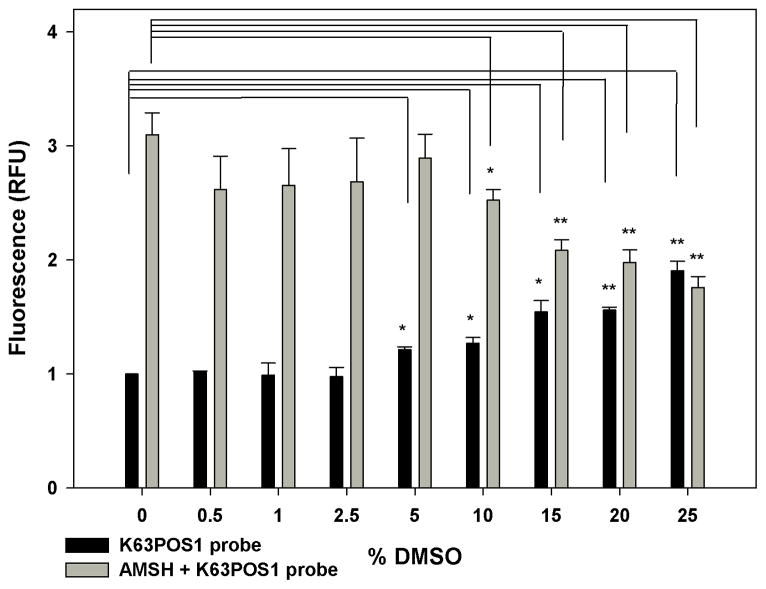
DMSO is a widely used solvent for small molecules, and commercially available chemical libraries used in screening campaigns are commonly shipped as DMSO solutions. Thus the effect of DMSO on the assay was assessed. Increasing concentrations of DMSO were titrated into a constant concentration of AMSH and incubated for 30 min. The K63POS1 probe was then added and the resulting mixture was incubated for an additional 90 min (Fig. 7). We observed a small but significant increase in the background at 5% DMSO with much larger effects at higher DMSO concentrations (Fig. 7, black bars). Additionally, a significant decrease in the signal at 10% DMSO (Fig. 7, grey bar) was observed. This suggests that DMSO has a significant effect on the assay, particularly as it relates to the screening window. Based on these observations, we conclude that the assay does not tolerate DMSO above 5% and during a HTS DMSO concentrations should be kept to a minimum and should not exceed 5%.
Figure 7.
Assay stability in the presence of increasing DMSO concentrations. Increasing concentrations of DMSO were added to AMSH (125 nM) and incubated 30 min before the addition of K63POS1 probe (500 nM). After a 90 min incubation with K63POS1 probe, fluorescence measurements were taken. (*p ≤ 0.035, **p ≤ 0.008). (n = 2)
Inhibition studies
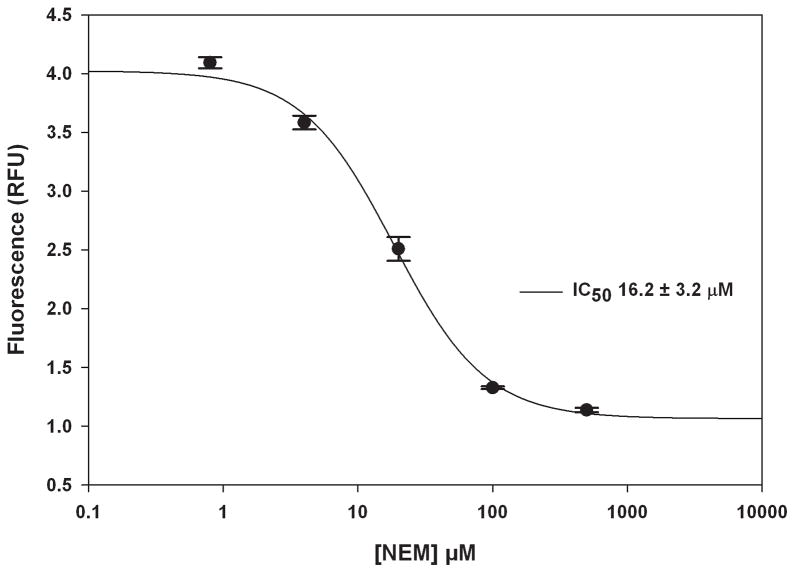
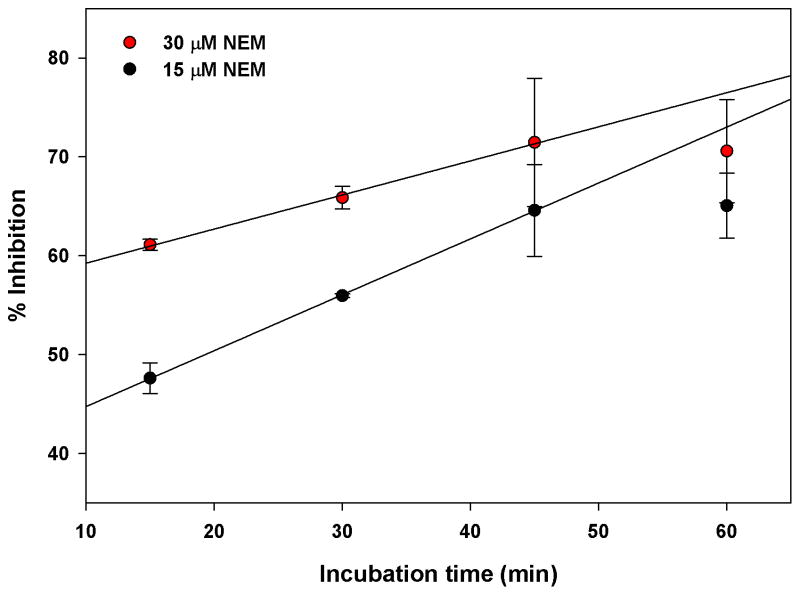
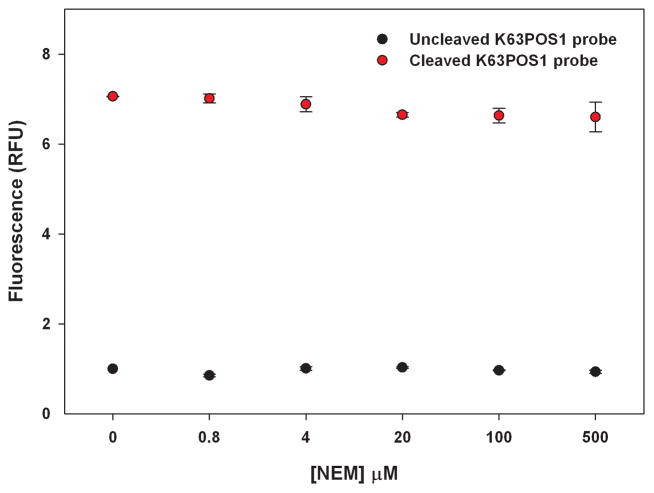
Currently there are no known selective inhibitors of AMSH however a positive control is desirable for a high-throughput screening campaign. A previous study showed that the nonselective Michael acceptor, N-ethylmalemide (NEM), inhibits AMSH, possibly by reacting with Cys282 near the active site [13]. Therefore to demonstrate that small molecules can inhibit AMSH in this FRET based assay format we determined the effects of NEM in the AMSH FRET assay (Fig. 8). AMSH was incubated with different concentrations of NEM (five-fold dilutions) for 30 min at room temperature. The K63POS1 probe was then added and the mixture incubated for an additional 90 min at 30°C. Fluorescence measurements revealed a dose dependent decrease with increasing concentrations of NEM indicating inhibition of AMSH (Fig. 8). The IC50 value (16.2 ± 3.1 μM) for NEM was determined by curve fitting the data. We also observed a time dependent effect of AMSH inhibition by NEM at 15 μM (~ IC50) and 30 μM (~ 2 × IC50) (Fig. 9). Increasing inhibition by NEM is observed up to 45 min of incubation with AMSH after which saturation is observed. This is likely due to all available NEM having reacted with AMSH at these concentrations. Additionally, to confirm that NEM was not affecting the FRET signal of the K63POS1 probe we incubated increasing concentrations of NEM (0–500 μM) with cleaved or uncleaved K63POS1 probe (Fig. 10). We observe no significant affects to the fluorescent signal in the presence of NEM. Since AMSH belongs to the JAMM family of DUB which are Zn metalloproteases, chelation agents phenanthroline and 2,2′-bypridine were also tested in this assay (data not shown). To our surprise, we observed inhibition of AMSH only at very high concentrations (> 1 mM) of the compounds. Based on these finding we suggest NEM as a qualitative positive control for high-throughput screening campaigns to identify reversible inhibitors. To identify hits from a HTS campaign, fluorescence values that are 3SD (99% confidence) from the mean of the negative control (DMSO) should be used and validated in subsequent dose response studies.
Figure 8.
Inhibition by N-ethylmaleimide. Five-fold dilutions of NEM were incubated with AMSH (125 nM) for 30 min at room temperature followed by the addition of the K63POS1 probe (500 nM). The plate was then read following a 90 min incubation at 30°C. (IC50 = 16.2 ± 3.1 μM). (n = 2)
Figure 9.
Time-dependent inhibition by N-ethylmaleimide. AMSH (125 nM) was incubated with either 15 μM or 30 μM NEM for 1 h, 45 min, 30 min or 15 min followed by the addition of the K63POS1 probe (500 nM). Fluorescent measurements were taken following a 90 min incubation at 30°C. (n = 2)
Figure 10.
Effect of N-ethylmaleimide on K63POS1 probe fluorescence. Increasing concentrations of NEM (0–500 μM) were incubated for 90 min at 30°C with cleaved or uncleaved K63POS1 probe before fluorescent measurements were taken. (n = 2)
Z-score
The suitability of an assay for high-throughput screening is established by the non-unit statistical parameter Z-score [31]. It is generally accepted that the Z-score reflects reproducibility, robustness, and reliability of an assay for HTS. Z-score for a given assay, range from 0 to 1, with 1 being ideal and a score of ≥ 0.5 is generally considered suitable for HTS. To determine the Z-score and inter-day and inter-plate variability for this assay, measurements were made with K63POS1 probe only, AMSH + K63POS1 probe, and AMSH + K63POS1 probe treated with 25 μM NEM for 120 min (Fig. 11) on separate days in different 384-well plates. The average Z-score was 0.71 indicating that the assay is reproducible, robust, and reliable and thus HTS compatible. Additionally, following a 24 h incubation at 4°C the plates were reread (data not shown). There was no significant difference in the overall Z-score of the plates, however there was increase in the fluorescence of AMSH + K63POS1 probe and AMSH + K63POS1 probe treated with NEM wells by ~9% and ~30% respectively. This increase in fluorescence is likely due to the cleavage of K63POS1 probe by AMSH over time even in the presence of inhibitor. Due to the potential variability in incubation times of plates during a high-throughput screen internal plates controls should be used to normalize for AMSH activity.
Figure 11.
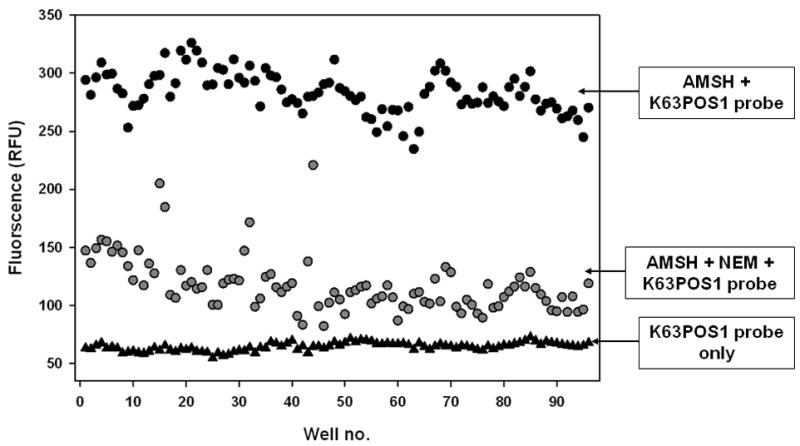
Z-score determination. K63POS1 probe was incubated either with buffer, AMSH, or AMSH + 25 μM NEM on separate days and plates. An overall Z-score of 0.71 was calculated. Well no. 1–48 are from day 1 and well no. 49–96 represent are from day 2. (n = 2)
In summary, using a FRET based system we showed that AMSH selectively cleaves Lys63-linked diubiquitin over Lys48- and Lys11-linked diubiquitin. The FRET AMSH assay was miniaturized to a 384-well format, with the optimal temperature for the screen determined to be 30°C and the maximum DMSO concentration tolerated was determined to be 5%. Further the Z-score for this assay was 0.71, which indicates HTS compatibility. Lastly, the reagents for this assay can be readily generated for a HTS with the major expense is the linkage specific FRET probe.
Supplementary Material
Acknowledgments
We would like to thank Elizabeth C. Blowers for editing the manuscript. Funding in part from Eppley Cancer Center, Nebraska Research Initiative and GAANN predoctoral fellowship (JLA). SMR acknowledges support from the Nebraska Center for Nanomedicine-Center for Biomedical Research Excellence (NCN-COBRE) seed grant. American Heart Association predoctoral fellowship to CWD and 1R01RR026273 to CD are gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wickliffe K, Williamson A, Jin L, Rape M. The multiple layers of ubiquitin-dependent cell cycle control. Chem Rev. 2009;109:1537. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science Signalling. 2007;315:201. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 5.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 6.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nature Reviews Molecular Cell Biology. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 7.Fraile J, Quesada V, Rodríguez D, Freije J, López-Otín C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2011;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncology. 2007;3:191–199. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka N, Kaneko K, Asao H, Kasai H, Endo Y, Fujita T, Takeshita T, Sugamura K. Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J Biol Chem. 1999;274:19129–19135. doi: 10.1074/jbc.274.27.19129. [DOI] [PubMed] [Google Scholar]
- 10.Clague MJ, Urbé S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Wright MH, Berlin I, Nash PD. Regulation of endocytic sorting by ESCRT–DUB-mediated deubiquitination. Cell Biochem Biophys. 2011;60:39–46. doi: 10.1007/s12013-011-9181-9. [DOI] [PubMed] [Google Scholar]
- 12.Nijman S, Luna-Vargas M, Velds A, Brummelkamp TR, Dirac AMG, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 13.McCullough J, Clague MJ, Urbé S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyuuma M, Kikuchi K, Kojima K, Sugawara Y, Sato M, Mano N, Goto J, Takeshita T, Yamamoto A, Sugamura K. AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct Funct. 2006;31:159–172. doi: 10.1247/csf.06023. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura M, Tanaka N, Kitamura N, Komada M. Clathrin anchors deubiquitinating enzymes, AMSH and AMSH- like protein, on early endosomes. Genes to Cells. 2006;11:593–606. doi: 10.1111/j.1365-2443.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- 16.Ma YM, Boucrot E, Villén J, Gygi SP, Göttlinger HG, Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J Biol Chem. 2007;282:9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 17.Meijer IMJ, van Rotterdam W, van Zoelen EJJ, van Leeuwen JEM. Recycling of EGFR and ErbB2 is associated with impaired Hrs tyrosine phosphorylation and decreased deubiquitination by AMSH. Cell Signal. 2012 doi: 10.1016/j.cellsig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Hasdemir B, Murphy JE, Cottrell GS, Bunnett NW. Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor 2. J Biol Chem. 2009;284:28453–28466. doi: 10.1074/jbc.M109.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrera-Vigenor F, Hernández-García R, Valadez-Sánchez M, Vázquez-Prado J, Reyes-Cruz G. AMSH regulates calcium-sensing receptor signaling through direct interactions. Biochem Biophys Res Commun. 2006;347:924–930. doi: 10.1016/j.bbrc.2006.06.169. [DOI] [PubMed] [Google Scholar]
- 20.Hislop JN, Henry AG, Marchese A, Von Zastrow M. Ubiquitination regulates proteolytic processing of G protein-coupled receptors after their sorting to lysosomes. J Biol Chem. 2009;284:19361–19370. doi: 10.1074/jbc.M109.001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reyes-Ibarra AP, García-Regalado A, Ramírez-Rangel I, Esparza-Silva AL, Valadez-Sánchez M, Vázquez-Prado J, Reyes-Cruz G. Calcium-sensing receptor endocytosis links extracellular calcium signaling to parathyroid hormone-related peptide secretion via a Rab11a-dependent and AMSH-sensitive mechanism. Molecular Endocrinology. 2007;21:1394–1407. doi: 10.1210/me.2006-0523. [DOI] [PubMed] [Google Scholar]
- 22.Sierra MI, Wright MH, Nash PD. AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J Biol Chem. 2010;285:13990–14004. doi: 10.1074/jbc.M109.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MS, Kim JA, Song HK, Jeon H. STAM–AMSH interaction facilitates the deubiquitination activity in the C-terminal AMSH. Biochem Biophys Res Commun. 2006;351:612–618. doi: 10.1016/j.bbrc.2006.10.068. [DOI] [PubMed] [Google Scholar]
- 24.Davies CW, Paul LN, Kim MI, Das C. Structural and Thermodynamic Comparison of the Catalytic Domain of AMSH and AMSH-LP: Nearly Identical Fold but Different Stability. J Mol Biol. 2011;413:416–429. doi: 10.1016/j.jmb.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCullough J, Row PE, Lorenzo Ó, Doherty M, Beynon R, Clague MJ, Urbé S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Current biology: CB. 2006;16:160. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 26.Inglese J, Johnson RL, Simeonov A, Xia M, Zheng W, Austin CP, Auld DS. High-throughput screening assays for the identification of chemical probes. Nature chemical biology. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 27.Goldenberg SJ, McDermott JL, Butt TR, Mattern MR, Nicholson B. Strategies for the identification of novel inhibitors of deubiquitinating enzymes. Biochem Soc Trans. 2008;36:828. doi: 10.1042/BST0360828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lokesh G, Rachamallu A, Kishore Kumar G, Natarajan A. High-throughput fluorescence polarization assay to identify small molecule inhibitors of BRCT domains of breast cancer gene 1. Anal Biochem. 2006;352:135–141. doi: 10.1016/j.ab.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 29.Kumar EA, Charvet CD, Lokesh G, Natarajan A. High-throughput fluorescence polarization assay to identify inhibitors of Cbl (TKB)–protein tyrosine kinase interactions. Anal Biochem. 2011;411:254–260. doi: 10.1016/j.ab.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simeonov A, Yasgar A, Jadhav A, Lokesh G, Klumpp C, Michael S, Austin CP, Natarajan A, Inglese J. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT–phosphoprotein interaction. Anal Biochem. 2008;375:60–70. doi: 10.1016/j.ab.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. Journal of Biomolecular Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.