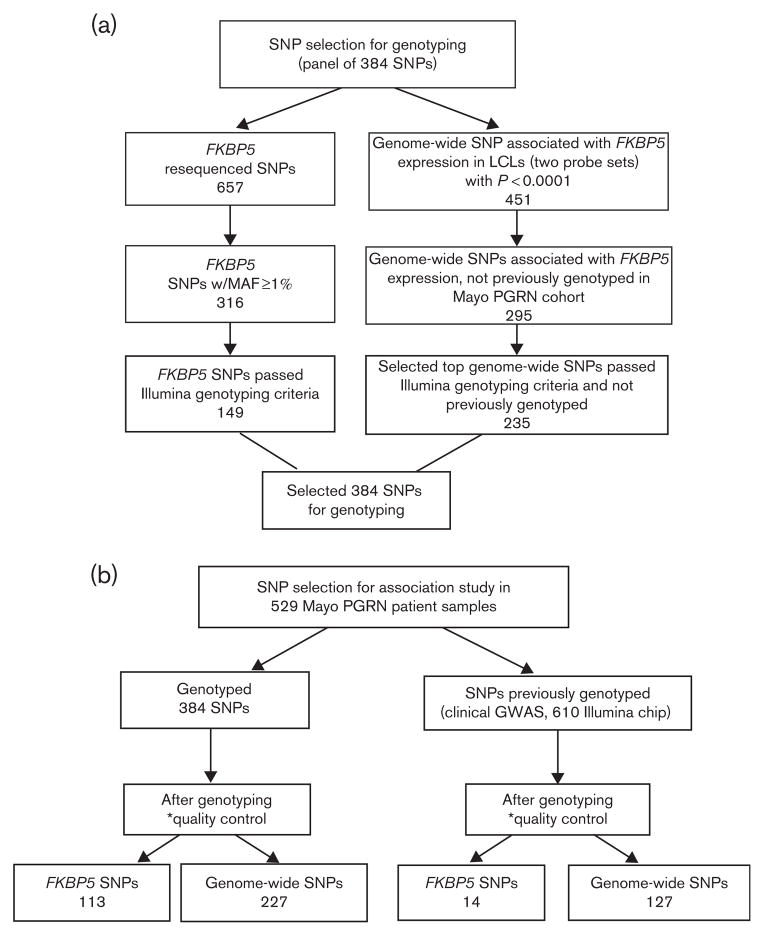
Fig. 1.
SNP selection for association analysis. The flow chart outlines the selection criteria for SNPs used for the genotyping and association studies. Genome-wide SNPs are FKBP5 eQTLs. *SNP level quality control. SNPs selected for an association analysis had to meet the quality control measures: minor allele frequency >0.01, per SNP call rate>0.95, and Hardy–Weinberg equilibrium P-value>0.001. eQTLs, expression Quantitative Trait Locus; GWAS, genome-wide association study; LCLs, lymphoblastoid cell lines; MAF, minor allele frequency; Mayo PGRN, Mayo Clinic Pharmacogenomics Research Network; SNPs, single nucleotide polymorphisms.