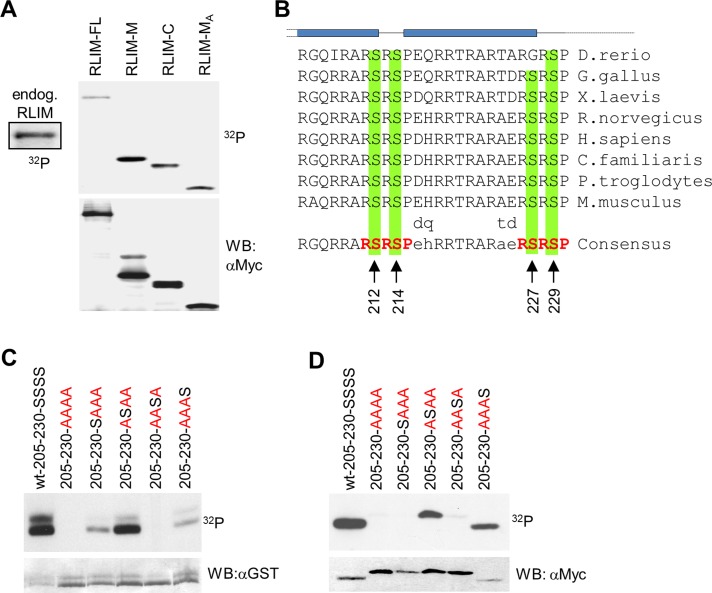
FIGURE 3:
The NLS of RLIM is phosphorylated at conserved Ser residues. (A) Metabolic labeling of transfected Myc-tagged RLIM protein mutants RLIM-M (amino acids [aa] 206–423), RLIM-C (aa 403–600), and RLIM-MA (aa 206–305) in cells. Note strong phosphorylation of RLIM-M and RLIM-MA deletion mutants, both containing the NLS. (B) Comparison of NLS of RLIM in vertebrates (205–230). Note the high conservation of the NLS. Serines at positions 212, 214, 227, and 229 that may serve as potential phosphorylation sites are indicated by arrows. RSRSP motifs are indicated in red. Predicted α-helices (GOR IV, PSIPRED, and Jnet) are indicated in blue. (C) In vitro phosphorylation of the mouse RLIM-NLS (205–230) fused to GST and RLIM-NLS Ser-to-Ala mutant proteins in which all serine residues were replaced by alanine residues (AAAA, in red) or containing only one serine residue at the indicated position. Note the strong serine phosphorylation at position 214. As loading control the same membrane was hybridized with an antibody directed against GST. (D) Metabolic labeling of Myc-tagged mouse RLIM-NLS (205–230) Ser-to-Ala mutations after transfection of corresponding expression plasmids. Note strong serine phosphorylation at positions 214 and 229. As a loading control the same membrane was hybridized with an antibody directed against Myc.