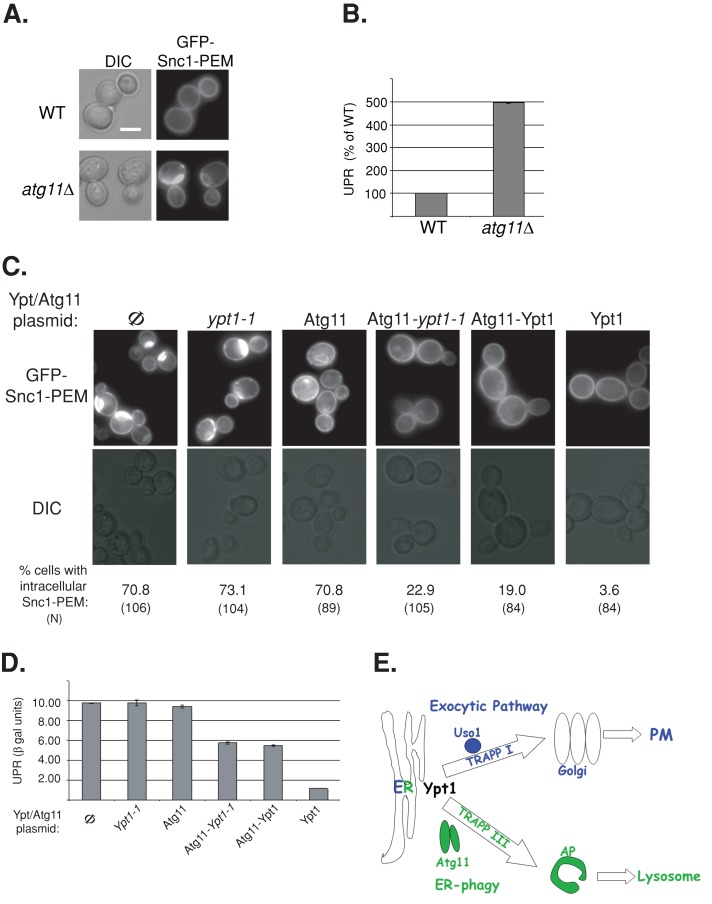
FIGURE 10:
A role for the Ypt1-Atg11 interaction in ER phagy. (A) GFP-Snc1-PEM accumulates in atg11∆ mutant cells. Wild-type and atg11∆ mutant cells expressing GFP-Snc1-PEM were visualized by live-cell microscopy. Representative cells are shown (25 cells for each strain), DIC (left) and GFP (right). (B) UPR is induced in atg11∆ mutant cells expressing GFP-Snc1. UPR was determined in wild-type and atg11∆ mutant cells expressing GFP-Snc1 and LacZ under a UPR-inducible promoter as in Figure 8. (C) The Atg11-Ypt1-T40K fusion protein complements the GFP-Snc1-PEM accumulation defect of ypt1-1 mutant cells. Ypt1-1 mutant cells expressing GFP-Snc1-PEM were transformed with a CEN plasmid to express the following proteins (left to right): Φ, empty vector control, Ypt1-1, Atg11, Atg11-Ypt1-T40K, Atg11-Ypt1, and Ypt1. Cells were visualized by live-cell microscopy, and representative cells are shown (GFP, top; DIC, bottom). Percentage of cells with intracellular GFP-Snc1-PEM is indicated at the bottom (N, number of cells visualized). Approximately 70% of ypt1-1 mutant cells accumulate intracellular GFP-Snc1-PEM (Φ, left). This defect can be suppressed by Ypt1 (right), as well as by the two fusion proteins, Atg11-Ypt1-T40K and Atg11-Ypt1, but not by Ypt1-T40K or Atg11 proteins. (D) Suppression of UPR induction in ypt1-1 mutant cells by the Atg11-Ypt1-T40K fusion protein. Ypt1-1 mutant cells were transformed with two plasmids expressing LacZ under a UPR-inducible promoter and Atg11/Ypt1. Induction of the UPR was determined using the βGAL assay. UPR is induced in ypt1‑1 mutant cells (Φ, left, and this induction is suppressed by overexpression of Ypt1 (right), as well as by the two fusion proteins, Atg11-Ypt1-T40K and Atg11-Ypt1, but not by Ypt1-T40K or Atg11 proteins. Bar, 5 μm; error bars, SD (B, D). Results shown in A–D represent at least two independent experiments. (E) Model depicting a role for Ypt1, together with its process-specific GEFs and effectors, in coordinating ER-to-Golgi and ER-phagy. Here we show that, whereas in wild-type cells some overexpressed GFP-tagged Snc1 is transported from the ER to the PM via the exocytic pathway, some is shuttled to the lysosome (vacuole in yeast) for degradation through the autophagic pathway. Ypt1 is required for both pathways. In the exocytic pathway Ypt1 is activated by TRAPP I and mediates ER-to-Golgi transport via vesicles; Uso1/p115 is an example of an ER-to-Golgi–specific effector (Cao et al., 1998; Allan et al., 2000). In autophagy, Ypt1 is activated by Trs85-containing TRAPP III to shuttle ER-derived membranes loaded with misfolded proteins to autophagosomes; Atg11 is an autophagy-specific effector (Lipatova et al., 2012). In ypt1-1, trs85∆, and atg11∆ mutant cells, shuttling of GFP-Snc1 from the ER to the autophagosome is defective, GFP-Snc1 accumulates in aberrant ER structures, and the ER-stress response, UPR, is induced.