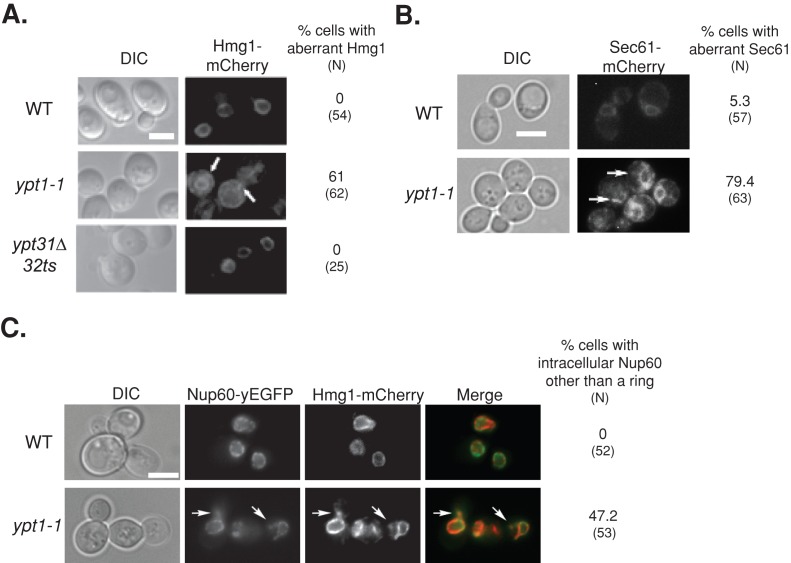
FIGURE 2:
Excess ER accumulates in ypt1-1 mutant cells expressing fluorescently tagged ER proteins. (A) Excess ER accumulates in ypt1-1 mutant cells expressing Hmg1-mCherry. The ER protein Hmg1 was tagged on the chromosome with mCherry in wild-type and ypt1‑1 and ypt31∆/32ts mutant cells. Cells were visualized by live-cell fluorescence microscopy. Left to right, differential interference contrast (DIC), mCherry, and percentage of cells with aberrant Hmg1 structures (N, number of cells visualized). In wild-type and ypt31∆/32ts mutant cells, mCherry-tagged Hmg1 localizes to rings around nuclei, a typical ER staining pattern (Huh et al., 2003). On the other hand, excess ER structures accumulate in the majority of ypt1‑1 mutant cells. (B) Excess ER accumulates in ypt1-1 mutant cells expressing Sec61-mCherry. Sec61 was tagged on the chromosome with mCherry in wild-type and ypt1‑1 mutant cells. Cells were visualized by live-cell fluorescence microscopy. Left to right, DIC, mCherry, and percentage of cells with aberrant Sec61 structures (N, number of cells visualized). In wild-type cells, mCherry-tagged Sec61 localizes to rings around nuclei and underneath the PM, as previously seen for Sec61-GFP (Huh et al., 2003). In addition, excess ER structures accumulate in the majority of ypt1‑1 mutant cells. (C) Excess ER accumulates in ypt1-1 mutant cells expressing Nup60–yeast enhanced GFP (yEGFP) and Hmg1-mCherry. The nuclear pore protein Nup60 was tagged on the chromosome with yEGFP in wild type and ypt1‑1 mutant cells expressing Hmg1-mCherry. Cells were visualized by live-cell fluorescence microscopy. Left to right, DIC, GFP, mCherry, merge, percentage of cells with Nup60 other than the ring (N, number of cells visualized). In wild-type cells, Nup60-GFP localizes to rings around nuclei (Huh et al., 2003). About half of the ypt1-1 mutant cells (47%) contain structures other than the rings in which Nup60-GFP colocalizes with Hmg1-mCherry (92%). Arrows point to aberrant ER structures; bars, 5 μm. Results represent at least two independent experiments.