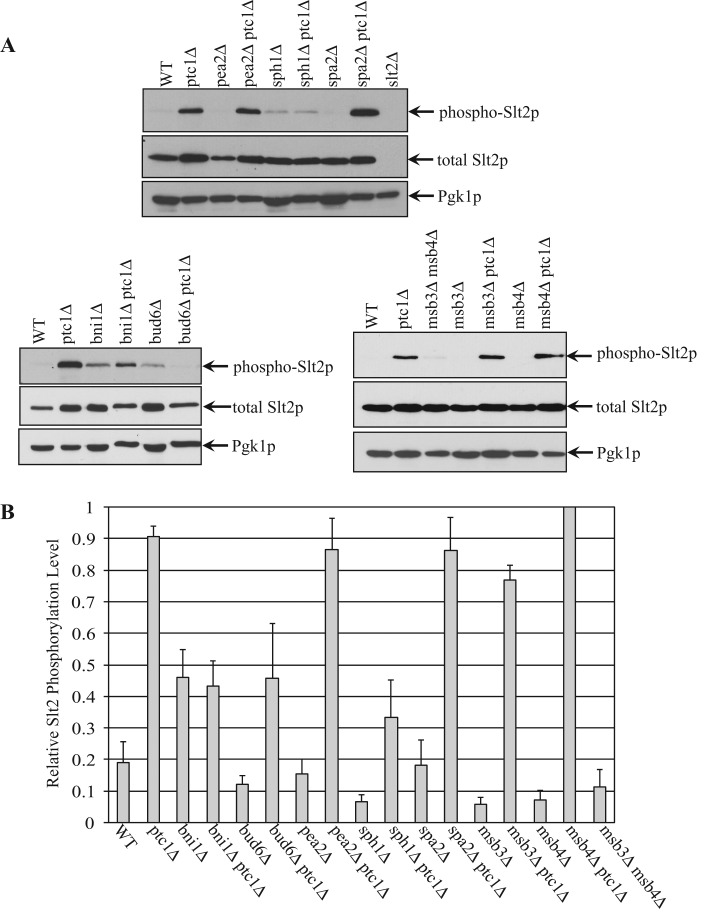
FIGURE 3:
Loss of Bni1p, Bud6p or Sph1p decreases the level of activated Slt2p in ptc1∆ mutant cells. (A) Yeast lysates were extracted separately from strains wt, ptc1∆, pea2∆, pea2∆ptc1∆, sph1∆, sph1∆ptc1∆, spa2∆, and spa2∆ptc1∆ (top), bni1∆, bni1∆ptc1∆, bud6∆, bud6∆ptc1∆ (bottom, left), or msb3∆msb4∆, msb3∆, msb3∆ptc1∆, msb4∆, and msb4∆ptc1∆ (bottom, right). Slt2p activation was measured as the intensity of dually phosphorylated Slt2p on Western blots, and the total Slt2p protein level in the same sample was measured using anti-Slt2p blots. Pgk1p was used as a loading control. (B) The relative phosphorylation level of Slt2p from the strains examined was quantified. The density of the phospho-Slt2p band was divided by the density of Pgk1p, and the normalized phospho-Slt2p value of each strain is shown in the bar graph. Error bars, SEM from four independent experiments.