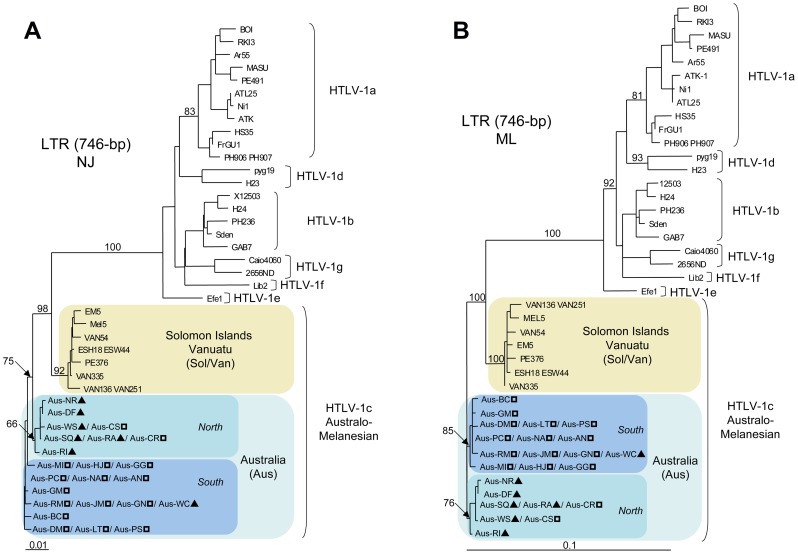
Figure 4. Phylogenetic trees generated with both Neighbor-Joining (figure 4A) and Maximum Likelihood (figure 4B) methods, performed in the PAUP program, on a 746-bp fragment of the LTR region, for 55 HTLV-1 available sequences including the 27 proviral sequences generated in this work.
The selected model was the General Time-Reversible (GTR). a-g correspond to the major HTLV-1 subtypes. HTLV-1c subtype corresponds to the Australo-Melanesian one including the 23 new Australian proviral sequences (Genbank: KC786899-KC786919 and JX891478-JX891479) and 4 sequences from Vanuatu (Genbank: KC786962-KC786965). ?, HTLV-1c subtype strains from Northern part of central Australia; ?, HTLV-1c subtype strains from Southern part of central Australia.