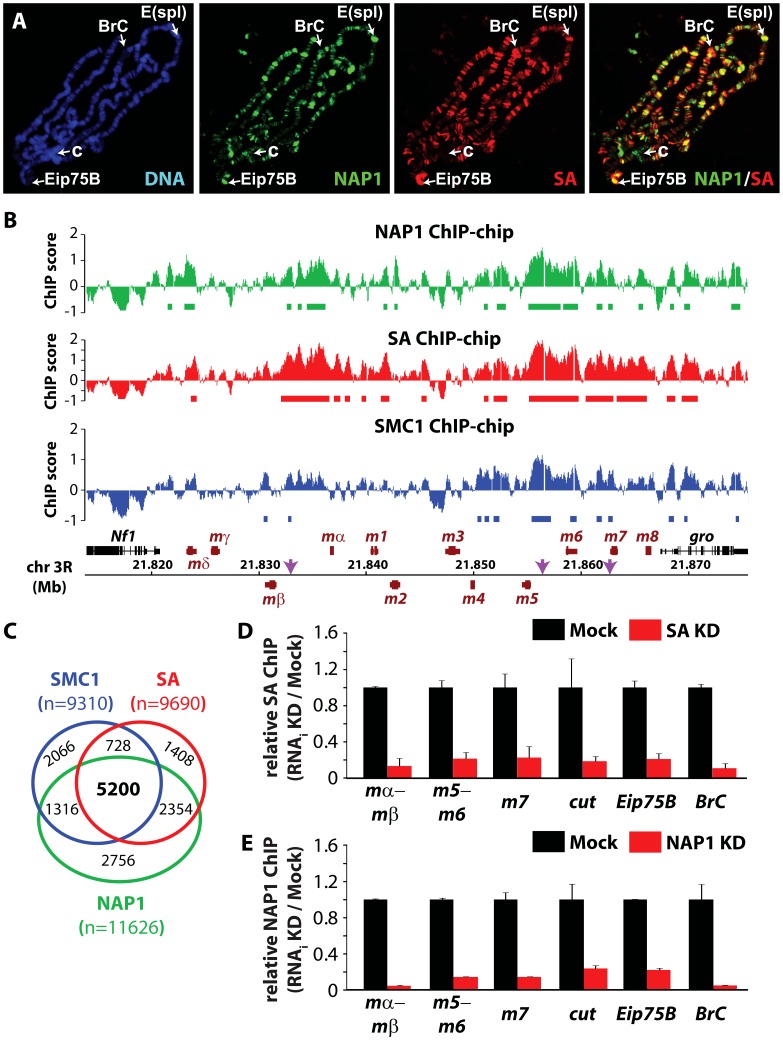
Figure 3. NAP1 and cohesin co-localize on chromatin.
(A) Distribution of NAP1 and SA proteins on Drosophila salivary gland polytene chromosomes visualized by indirect immunofluorescence with antibodies against NAP1 (green) and SA (red). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Split images and merge for red and green channels are shown. Regions harboring the Enhancer of split (E(spl)) gene cluster, Broad Complex (BrC), Eip75B and chromocenter (c) are indicated. (B) Genomic view of NAP1 (green), SA (red) and SMC1 (blue) ChIP-chip enrichment profiles at Enhancer of Split E(spl) NOTCH inducible gene cluster. Filtered binding sites (FDR<0.01) are indicated as bars below the respective profiles. ChIP-chip enrichment scores, genomic coordinates and genes are indicated. Regions examined by ChIP-qPCR are indicated by arrows. (C) Venn diagram depicting the overlap between SMC1, SA and NAP1 binding loci. The overlap between SMC1 and SA loci is 3.7 times greater (p<0.001) than expected by random chance. The overlap between NAP1 and cohesin loci derived from shared SMC1 and SA filtered peaks is 5.2 times greater (p<0.001) than expected by random chance. (D) ChIP-qPCR analysis of SA binding to genomic sites harboring E(spl), cut, Eip75B and BrC genes selected from ChIP-chip profiles (Figure 3B and Figure S3A–C). ChIP enrichments after SA (red bars) and Mock (black bars) RNAi knockdowns (KD) were expressed relative to signals from mock-treated cells. For mock treatment we used dsRNAs directed against GFP. Results are based on 3 biological replicates and error bars represent standard error of mean (S.E.M.). (E) ChIP-qPCR analysis of NAP1 binding to genomic loci. Analysis as described above.