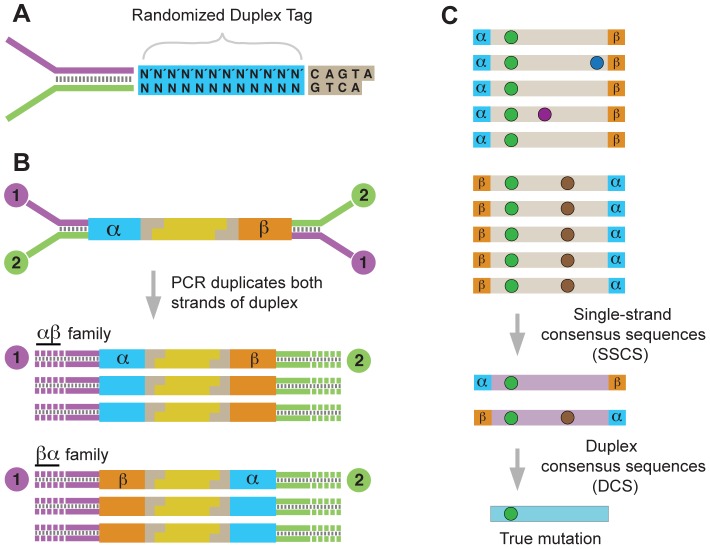
Figure 1. Overview of the Duplex Sequencing methodology.
(A) Adapter design with random double-stranded tag sequence and invariant spacer sequence. (B) Ligation of adapters to fragmented DNA generates unique 12 bp tags on each end (α and β). PCR amplification of the two strands produces two related, but distinct products. (C) Sequence reads sharing unique α and β tags are grouped into families of α-β or β-α orientation. Mutations are of three different types: sequencing mistakes or late arising PCR errors (blue or purple spots); first round PCR errors (brown spots); true mutations (green spots). Comparing SSCSs from the paired families generates a DCS, which eliminates all but true mutations.