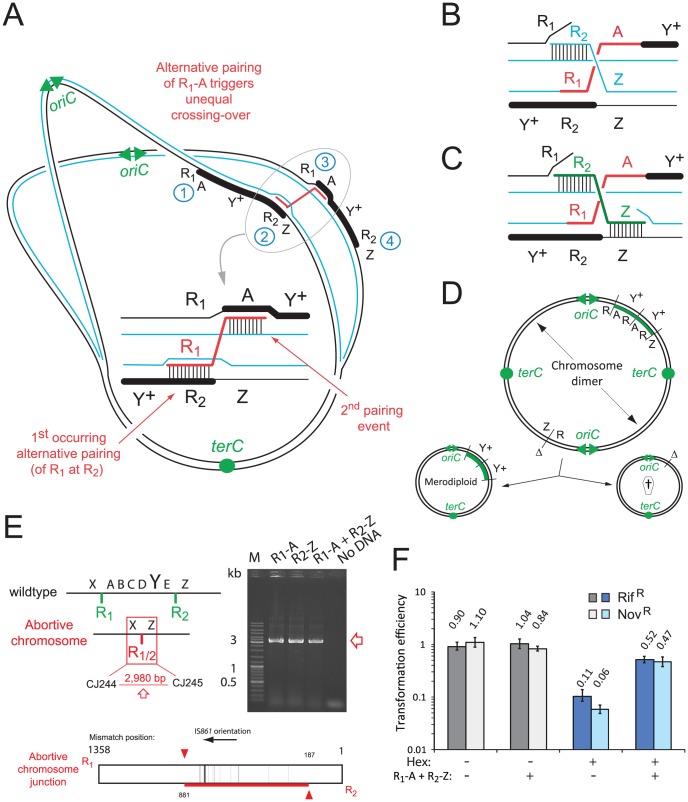
Figure 5. Model of merodiploid formation during pneumococcal transformation.
(A) Alternative pairing between R-NRf sequences triggers unequal crossing over. Letters as in Figure 1D. (B) Spontaneous restoration of complementary strand integrity. (C) Restoration of complementary strand integrity stimulated by R2-Z donor fragment. (D) Resolution of chromosome dimer to produce one tandem-duplicated chromosome and one abortive chromosome. oriC, origin of replication; terC, terminus of replication; Δ, deletion; †, abortive chromosome. (E) Predicted abortive chromosome structure. Layout as in Figure 2A. Red square; abortive chromosome junction. Detection of abortive chromosome junction by primers in Figure 3C. Diagram of R1/2 sequence at abortive chromosome junction amplified after R1-A transformation, determined by sequencing of PCR fragments. Vertical lines, R1/R2 mismatches; red horizontal line, mixed sequence .(F) Saturation of the Hex system establishes frequent alternative pairing of R-NRf fragment. Transformation efficiencies of RifR and NovR point mutations (carried by R304 chromosomal DNA) compared in hex− (R246) or hex+ (R800) recipients in the presence or absence of R-NRf PCR fragments. Transformation efficiencies normalized to SmR (rpsL41) point mutation, resistant to Hex. hex − recipient, shades of gray; hex + recipient, shades of blue.