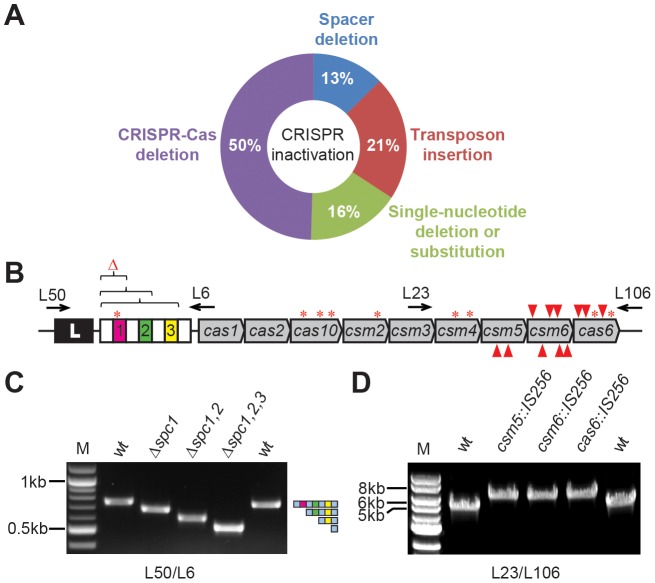
Figure 2. Different mutations eliminate CRISPR immunity against conjugation in S. epidermidis.
(A) Summary of the different mutations found in this study and their proportions. (B) Distribution of mutations within the CRISPR-Cas locus. S. epidermidis RP62a harbors a CRISPR-Cas system containing four repeats (white boxes), three spacers (colored, numbered boxes) and nine cas/csm genes. Mutations found in CRISPR escapers include deletions in the repeat-spacer region (brackets), transposon insertions (red arrowheads; top, direct insertion; bottom, inverted) and single nucleotide deletions or substitutions (asterisks). Arrows indicate primers used to analyze transconjugants. (C) PCR analysis of the CRISPR array of transconjugants using primers L50/L6. Deletion of 1, 2 and 3 spacers observed in escapers R23, R10 and R2, respectively, is shown. M, DNA marker. wt, amplification using wild-type template DNA. (D) PCR analysis of the cas gene region of escapers using primers L23/L106. IS256 transposon insertions into csm5, csm6 and cas6 observed in escapers R60, B15 and R36, respectively, are shown. M, DNA marker. wt, amplification using wild-type template DNA.