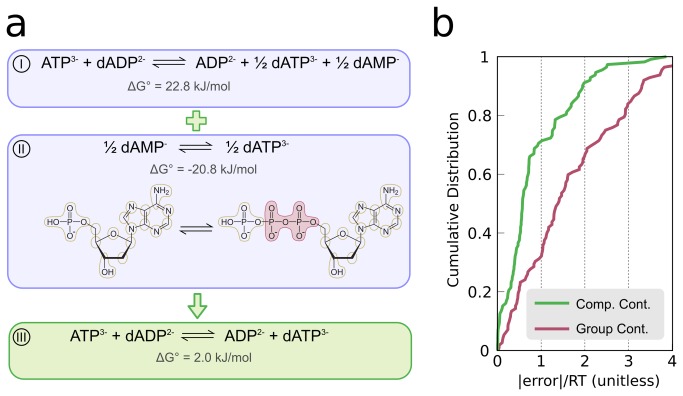
Figure 5. Using CCM to evaluate a reaction Gibbs energy.
(a) The reaction catalyzed by phosphoribosyl formyl glycinamidine synthase (I) involves mostly compounds from TECRDB and two others: N2-Formyl-N1-(5-phospho-D-ribosyl) glycinamide (FGAM) and 2-(Formamido)-N1-(5-phospho-D-ribosyl) acetamidine (FPRAM). In this case, CCM divides the reaction into two parts: one which can be completely evaluated using directly observed reaction data (II) and another which can only be resolved using group contributions (III). The five groups that change throughout this reaction are highlighted in red. The final ΔG'0 estimation is simply the sum of these two half-reactions: -164.0 + 204.9 = 40.9 kJ/mol. The last stage in the algorithm is to apply the Legendre transform [59] (using pKa data from ChemAxon; see Supp. Material). The result in this case is ΔG'0= -39.8 kJ/mol. The previous GCM prediction for the same reaction is ΔG'0= -90.8 kJ/mol (as appears in iAF1260). Note that the values appearing in iAF1260 are also Legendre transformed. (b) The cumulative distribution functions of the absolute errors for CCM and versus that of GCM taken from the iAF1260 model. The error was calculated by comparing the prediction to the median value for that reaction taken from TECRDB. Only reactions that appear in all three datasets are shown (113 in total). The intersections with the dashed lines indicate the fraction of reactions whose predicted value are in the range of ±RT (2.5 kJ/mol) of the value in TECRDB. One can see that for CCM, 70% of these reactions are in this category, compared to only about 30% in iAF1260.