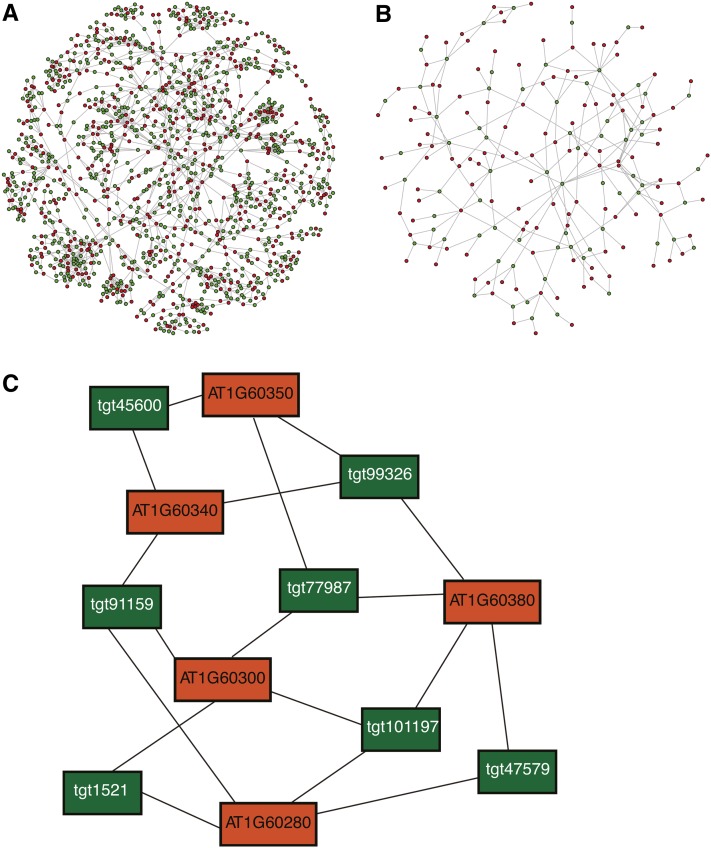
Figure 3.
Visualization of Relationships between AmiRNAs (Green) and Targeted Genes (Red).
(A) The network shown visualizes the largest connected unit in the PHANTOM library. The library assembly procedure maximizes the number of unique groups of targeted genes. Thus, the connectivity and complexity of the network is high. The target genes are part of the Kinomer TKL (Tyr kinase-like kinases) family, by far the largest group of kinases in land plants (Martin et al., 2009). AmiRNAs (green circles, nodes) are connected by edges (gray lines) to their target genes (red circles, nodes). Green circles represent one amiRNA sequence.
(B) The network shown visualizes the connected unit in an alternative assembly procedure, which was tested for an initial PHANTOM library design but not pursued further (see Results). This alternative assembly procedure maximizes the number of amiRNAs with a minimal set of groups of targeted genes. This reduces the connectivity and complexity of the network. The target genes are part of the AP2 transcription factor family (Kim et al., 2006). AmiRNAs (green circles, nodes) are connected by edges (gray lines) to their target genes (red circles, nodes). Green circles represent here more than one amiRNA sequence.
(C) Example of a typical target class target gene network in the PHANTOM amiRNA library representing members of the NAC transcription factor family. Orange rectangles represent target genes, while green rectangles represent amiRNAs labeled by an identifier. Note that most gene pairs are targeted by two or more amiRNAs, and each amiRNA targets a separate group of genes, thus enhancing robustness and gene combinations for screening.