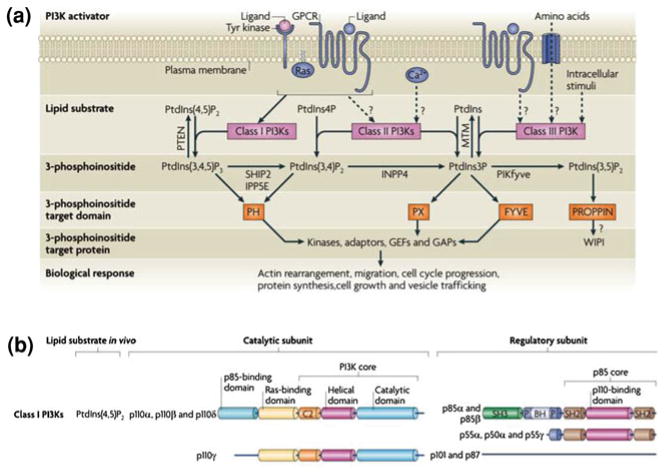
Fig. 3.
The 3-phosphoinositide lipid network. (a) Following activation by upstream agonists, phosphoinositide 3-kinases (PI3Ks) generate phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3), PtdIns-3,4-bisphosphate (PtdIns(3,4)P2) and PtdIns-3-phosphate (PtdIns3P). These lipids interact with lipid binding domains in PI3K effector proteins and change their localization and/or activity. Lipid phosphatases degrade or interconvert 3-phosphoinositides. These include lipid phosphatases for PtdIns(3,4,5)P3, such as phosphatase and tensin homologue deleted on chromosome 10 (PTEN), inositol polyphosphate-5-phosphatase E (IPP5E), and SH2 domain-containing inositol 5-phosphatase type 2 (SHIP2). GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GPCR, G protein-coupled receptor. (b) Classification and domain structure of mammalian Class I PI3Ks. All PI3K catalytic subunits have a PI3K core structure consisting of a C2 domain, a helical domain and a catalytic domain. Class I PI3Ks exist in complex with a regulatory subunit, either a p85 isoform (for p110α, p110β and p110δ) or p101 or p87 (for p110γ). All p85 isoforms have two Src homology 2 (SH2) domains and are encoded by either PIK3R1 (which encodes p85α, p55α and p50α), PIK3R2 (which encodes p85β) and PIK3R3 (which encodes p55γ). Figure modified from (Vanhaesebroeck et al. 2010)