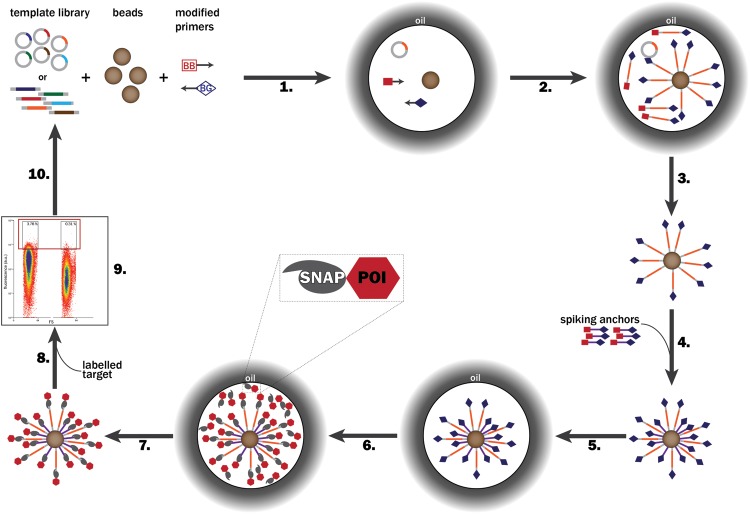
Fig. 2.
Steps of a directed evolution cycle using BeSD. (1) A DNA library (coding for SNAP-tag-fused POI variants), streptavidin-coated beads and the PCR mix containing BB forward primers and BG-labeled reverse primers are compartmentalized in water-in-oil emulsion droplets so that each compartment contains no more than one DNA template and one bead. (2) DNA is amplified by ePCR and captured on the beads via a biotin–streptavidin linkage. (3) The emulsion is broken, beads washed (to remove PCR mix components and unbound DNA that would compromise IVTT efficiency) and (4) spiking anchors added to provide extra display functionalities. (5) A new emulsion is then created in the presence of an IVTT system. (6) Individual SNAP-tagged POI variants are expressed in vitro and covalently linked to the BG-modified template DNA and spiking anchors via the SNAP-tag. (7) After de-emulsification and washing the beads are recovered. (9) The beads are incubated with the labeled target. (9) The affinity for the target is measured by FACS and the selected beads are isolated. (10) The identity of the selected clones is decoded after single-bead PCR by sequencing. Alternatively, the selected beads are used directly for another evolution cycle. The binding affinity of each recovered variant can be measured by subsequent FACS analysis on the bead display construct (see Fig. 6b). POI, protein or peptide of interest.