Abstract
Background
Most cells possess a sophisticated mechanism for sensing glucose and responsing to it appropriately. Glucose sensing and signaling in the budding yeast Saccharomyces cerevisiae represents an important paradigm for understanding how extracellular signals lead to changes in the gene expression program in eukaryotes.
Scope of review
This review focuses on the yeast glucose sensing and signaling pathways that operate in a highly regulated and cooperative manner to bring about glucose-induction of HXT gene expression.
Major conclusions
The yeast cells possess a family of glucose transporters (HXTs), with different kinetic properties. They employ three major glucose signaling pathways— Rgt2/Snf3, AMPK, and cAMP-PKA—to express only those transporters best suited for the amounts of glucose available. We discuss the current understanding of how these pathways are integrated into a regulatory network to ensure efficient uptake and utilization of glucose.
General significance
Elucidating the role of multiple glucose signals and pathways involved in glucose uptake and metabolism in yeast may reveal the molecular basis of glucose homeostasis in humans, especially under pathological conditions, such as hyperglycemia in diabetics and the elevated rate of glycolysis observed in many solid tumors.
Keywords: Glucose uptake and metabolism, glucose transporters, glucose signaling pathways, yeast, cancer
1. Introduction
Glucose serves as a metabolic substrate as well as a signaling molecule that regulates physiological and pathological processes [1-5]. Mammals maintain a constant level of glucose in the bloodstream despite intermittent supplies from the gut, and impaired regulation of blood glucose levels causes severe disorders such as diabetes [5,6]. Metastasized tumor cells metabolize large amounts of glucose through glycolysis and produce copious amounts of lactic acid even in the presence of oxygen [7,8], called the Warburg effect, a hallmark of cancer [9]. The well-established elevated glucose consumption of malignant tissue forms the basis of the clinical imaging of cancer, PET (positron emission tomography) [10]. Therefore, normal cells sense glucose levels in their environment and adapt central metabolic pathways to glucose availability [5]. Dysregulation of these pathways likely induces the glycolytic phenotype—increased glycolysis—in cancer cells [7,8].
Glucose is by far the preferred energy source of the budding yeast S. cerevisiae, because glucose regulation dictates the organism’s distinctive fermentative lifestyle—aerobic ethanol fermentation (the Crabtree effect) [11,12]. This is mediated, in part, by the crosstalk between the two glucose signaling pathways: 1) the Rgt2/Snf3 glucose induction pathway responsible for glucose uptake [1,13,14]; 2) the Snf1/Mig1 glucose repression pathway that negatively regulates the genes involved in the glucose oxidation and the use of alternative sugars [15-17]. Because the generation of the glucose repression signal that inhibits the activity of the Snf1 kinase (the yeast homolog of AMPK) requires glucose uptake and metabolism [18], yeast cells likely lock the signaling pathways in a cross talking network to determine its sensitivity to environmental changes in glucose availability [3,19,20]. The mechanistic basis of the Warburg effect in tumor cells, though not well understood, shows some remarkable similarities to the mechanism responsible for this phenomenon in yeast [14,21].
Since energy generation by fermentation is inefficient, yeast cells pump a large amount of glucose through glycolysis by enhancing the first, rate-limiting step of glucose metabolism—its uptake [1,22]. They do so by increasing expression of glucose transporter (HXT) genes. S. cerevisiae possesses at least six members of glucose transporter family (Hxt1, 2, 3, 4, 6, and 7), each with different affinities for glucose in order to cope with environmental changes in glucose availability [22,23]. The yeast cells have evolved mechanisms for sensing glucose over a broad concentration range and respond to it promptly and appropriately [3,19,20,24]. They express only those glucose transporters best suited for the amount of glucose available in the environment: the low affinity glucose transporter Hxt1 is only expressed when glucose levels are high (> ∼1%); in contrast, the high-affinity glucose transporters Hxt2 and Hxt4 are expressed when glucose levels are low (∼0.2%); the intermediate-affinity glucose transporter Hxt3 is expressed in both low and high glucose concentrations [1,25]. Expression of the high affinity glucose transporters Hxt6 and Hxt7 is highly induced under low glucose conditions or in the presence of non-fermentable carbon sources such as glycerol or ethanol, but is strongly repressed by high concentrations of glucose [26-28]. In addition, Hxt6 and Hxt7 are subject to glucose-induced degradation in the vacuole [29]. In this review, we focus on the crosstalk between the glucose signaling pathways that leads to induction of HXT gene expression in yeast, as a model to study how cells sense glucose and adapt their gene expression program to glucose availability.
2. Key components of the HXT gene repressor complex
Rgt1 binds to the upstream regulatory region of the HXT genes in the absence of glucose and brings about repression by recruiting the general repressor complex Ssn6-Tup1. Rgt1 does so in conjunction with the glucose responsive transcription factors Mth1 and Std1. These components form a repressor complex that inhibits transcription of HXT genes in the absence of glucose. A unique feature of Rgt1-mediated repression is that, unlike other repressors, Rgt1 cannot recruit Ssn6-Tup1 directly; the interaction between Rgt1 and Ssn6-Tup1 requires Mth1 (Fig. 1). The respective roles of the three components—Rgt1, Mth1/Std1, and Ssn6-Tup1—in the formation of this complex are discussed in the context of a current model of Rgt1-mediated repression. Rgt1 recognizes a consensus sequence of 5’-CGGANNA-3’ (N is any nucleotide) and binds to multiple copies of the DNA sequence in a nonadditive, synergistic manner [30,31]. Functional studies of synthetic, multimerized Rgt1-binding sites without intervening sequences show that reporter genes containing 2 or 3 Rgt1-binding sites provide 2- to 3-fold repression and that those containing 4 and 6 such sites bring about 30- and 90-fold repression, respectively [30-32].
Fig. 1.
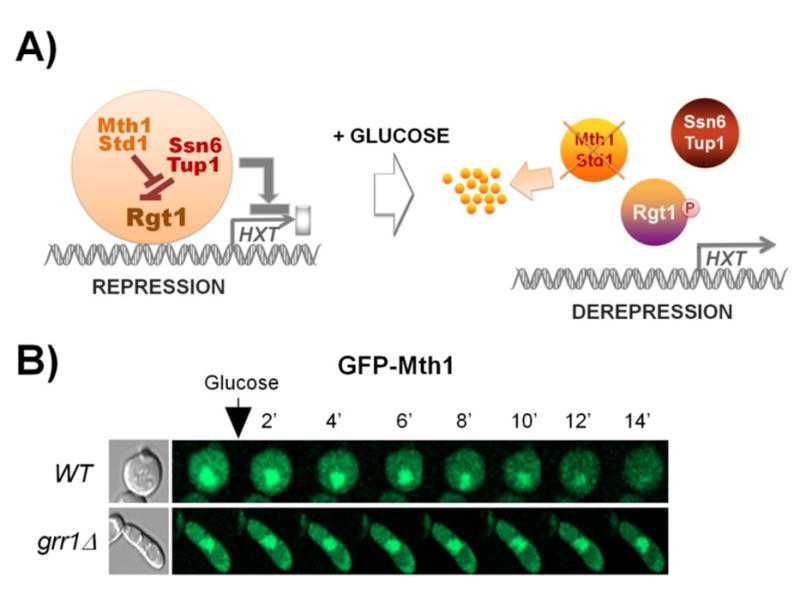
Mth1 is required for the interaction of Rgt1 with Ssn6-Tup1 that leads to repression of HXT gene expression; indeed, its inactivation is critical for glucose uptake and metabolism. A) Rgt1 recruits Ssn6-Tup1 in an Mth1-dependent manner and brings about repression of its target genes, such as HXT and HXK2 genes. Std1 is a paralog of Mth1 but has little effect on regulating Rgt1 function. Glucose-induction of HXT gene expression is achieved by a two-step process: (1) Mth1 and Std1 are degraded by the ubiquitin-proteasome pathway, rendering the PKA phosphorylation sites in Rgt1 available for phosphorylation; (2) Rgt1 phosphorylation by PKA induces its dissociation from Ssn6-Tup1 and consequently from HXT promoters. B) Time-lapse observation of Mth1 degradation in high-glucose medium (4%). Mth1 and Std1 are ubiquitinated by the SCFGrr1 ubiquitin-ligase, and the ensuing ubiquitination of Mth1 and Std1 targets them to the proteasome for degradation. The figure was adapted from [45].
2.1, The Rgt1 repressor
Rgt1 is a member of the Gal4 family of transcription factors that contains the zinc binuclear cluster (Cys6Zn2) DNA-binding domain [33]. Most members of the family bind as dimers to two ‘CGG’ triplets, whereas Rgt1 lacks the coiled-coil dimerization domain and thus binds DNA as a monomer [30]. This synergistic repression is probably due to efficient recruitment of Rgt1 to multiple binding sites [32]. Thus, Rgt1, as a monomer, functions more efficiently through its multiple sites. This is reminiscent of the Aspergillus nidulans transcriptional activator AlcR, which binds DNA as a monomer but functions synergistically through multiple sites [34]. Rgt1 is dissociated from the HXT promoters within 10 min after addition of glucose to glucose-depleted cultures. Rgt1 is phosphorylated at a basal level in the absence of glucose and hyperphosphorylated by PKA in high levels of glucose. Hyperphosphorylated Rgt1 does not bind DNA, whereas dephosphorylation of Rgt1 in vitro restores its DNA-binding ability [30,35,36]. Accordingly, the inhibition of Rgt1 phosphorylation is critical for the formation of the repressor complex (Fig. 1A).
2.2, The glucose responsive transcription factors Mth1 and Std1
Mth1 and Std1 are paralogous proteins that play a key role in regulation of Rgt1 function [37,38]. The main lines of supporting evidence are: 1) HXT gene expression is constitutive in the absence of Mth1 and Std1 [37-40] ; 2) Rgt1 does not bind to HXT promoters in yeast cells lacking both MTH1 and STD1 genes [35,36] ; 3) Mth1 and Std1 directly interact with Rgt1 [40-42]. Since Rgt1 is hyperphosphorylated in the mth1std1 mutant, Mth1 and Std1 are thought to serve as Rgt1 regulators that modulate the phosphorylation state of Rgt1 [36]. Mth1 and Std1 are downregulated by glucose; they are ubiquitinated by the SCFGrr1 ubiquitin ligase complex and degraded via the 26S proteasome in response to glucose [35,43-45] (Fig. 1B). There are also dominant mutations in the MTH1 gene (HTR1-23, DGT1 or BCP1[46,47]) that render Mth1 resistant to glucose-induced degradation [48]. Accumulating evidence shows that Mth1 and Std1 may be not functionally redundant. Deletion of the STD1 gene alone has little effect on the phosphorylation [42] and DNA-binding of the Rgt1 repressor [35] and the expression of HXT genes [48]. Furthermore, transcriptome analysis shows that expression of major HXT genes is profoundly upregulated in an mth1 mutant but is not significantly changed in a std1 mutant [49]. Therefore, Mth1 but not Std1 seems to be the major regulator of Rgt1, and the role of Mth1 in Rgt1-mediated repression is to block PKA-dependent Rgt1 phosphorylation in the absence of glucose.
2.3, The general corepressor complex Ssn6-Tup1
The Ssn6-Tup1 complex is a general transcriptional corepressor complex, composed of one molecule of Ssn6 and four molecules of Tup1 [50]. The complex contains the tetratrico peptide repeat (TPR) and WD domains, respectively, which serve as protein-protein interaction motifs [51-55]. Ssn6-Tup1 is recruited to its target promoters by sequence-specific DNA-binding repressors [56,57] and mediates transcriptional repression by recruiting global corepressors such as chromatin and nucleosome remodelers [58,59] and/or by interacting with the RNA transcription machinery [56,57]. Ssn6-Tup1 also appears to be involved in the induction of gene expression [60-62] and recruited to its target promoters in a manner independent of sequence-specific DNA-binding proteins [62-65]. In addition, Ssn6-Tup1 is shown to exert its function by masking the activation domain of a DNA-binding repressor and thereby preventing recruitment of the coactivators necessary for transcriptional activation [66].
Rgt1-mediated repression requires the Ssn6-Tup1 complex [33]. Rgt1 interacts with Ssn6 via the N-terminal domain (aa 210-250), located close to the Zn cluster DNA-binding motif [42]. However, recent studies show that Ssn6-Tup1 negatively regulates the DNA-binding ability of Rgt1. It does so in two ways: (1) by directly interfering with Rgt1 DNA-binding [67]; (2) by repressing expression of the MTH1 gene via the Snf1-Mig1 pathway [13]. The inhibitory function of Ssn6-Tup1 on Rgt1 is antagonized by Mth1 but becomes effective when Mth1 is degraded in high glucose conditions. The interaction of Rgt1 with Ssn6-Tup1 is unstable and readily dissociated in the absence of Mth1, establishing the role of Mth1 in Rgt1-Ssn6-Tup1 interaction [67] (Fig. 1A).
2.4, Ssn6-Tup1 regulates glucose induction and repression of gene expression
The finding that Ssn6-Tup1, although required for Rgt1-mediated repression, acts to inhibit Rgt1 function provides a new concept of how Ssn6-Tup1 can be appropriately targeted by distinct DNA-binding repressors [67]. Although the biological significance of this phenomenon is not fully understood, it can be related to the differential regulation of Ssn6-Tup1 target genes in response to glucose (Fig. 2). In high glucose conditions, the glucose repressor Mig1 recruits Ssn6-Tup1 to bring about repression of its target genes [68,69]; in contrast, Rgt1 is dissociated from Ssn6-Tup1, leading to derepression of Rgt1-repressed genes. Ssn6-Tup1 is actively involved in preventing the interaction with Rgt1 not only by repressing expression of the MTH1 gene but also by interfering with the DNA-binding ability of Rgt1 (Fig. 2A and see below). As a result, Ssn6-Tup1 does not interact with Rgt1 while in association with Mig1 in high glucose conditions, leading to highly regulated control of the glucose induction and repression of glucose-regulated genes. Thus, the two glucose responsive repressors Rgt1 and Mig1 are regulated in a similar manner [17,70]. Ssn6-Tup1 is recruited to unphosphorylated Mig1, but not to phosphorylated Mig1 (Fig. 2B). Similarly, the Rgt1-Ssn6-Tup1 interaction is disrupted when Rgt1 is hyperphosphorylated. Therefore, Mig1 and Rgt1 employ phosphorylation as a mechanism to regulate their interaction with Ssn6-Tup1.
Fig. 2.
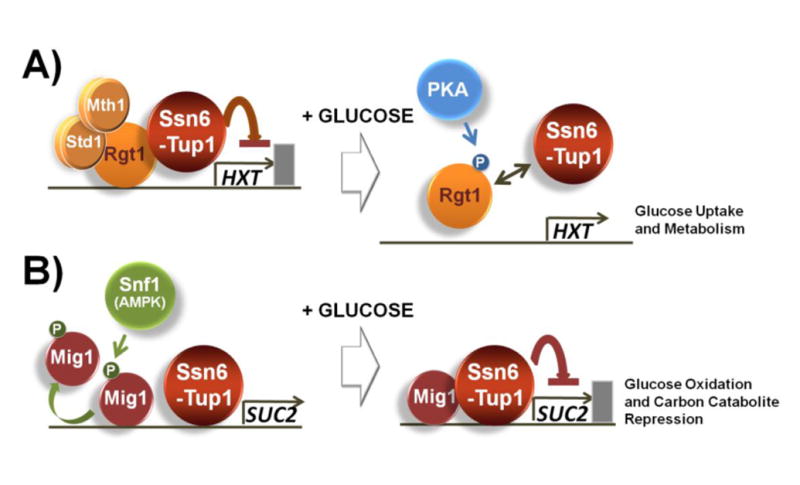
The two glucose responsive repressors Rgt1 and Mig1 are regulated in a similar manner. A) Rgt1 recruits Ssn6-Tup1 in the absence of glucose; however, it is hyperphosphorylated by PKA in the presence of high levels of glucose and dissociated from Ssn6-Tup1, resulting in the induction of expression of genes involved in glucose uptake and metabolism. B) Ssn6-Tup1 interacts with only unphosphorylated Mig1 in high levels of glucose and mediates the repression of genes involved in glucose oxidation and carbon catabolite repression. Snf1-dependent phosphorylation of Mig1 in glucose-limited conditions abolishes interaction with Ssn6-Tup1 [62].
3. Mechanism of glucose-induction of HXT gene expression
Glucose induces expression of the HXT genes by inhibiting the function of Mth1 and Rgt1. Mth1 mRNA and protein levels are down-regulated by glucose via the aforementioned glucose signaling pathways. Rgt1 is phosphorylated by the third glucose signaling pathway, the cAMP-activated kinase (PKA) pathway, which occurs when Mth1 is removed. Thus, three glucose signaling pathways converge at multiple points to induce expression of the HXT genes (Fig. 3).
Fig. 3.
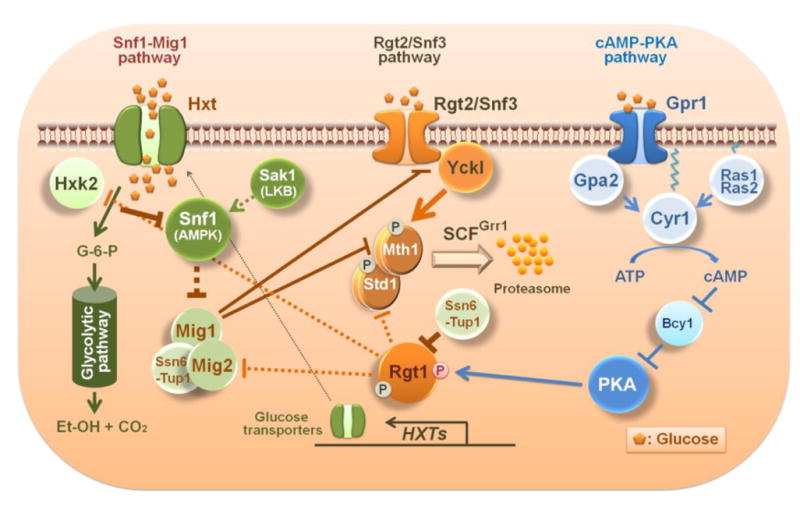
Schematic diagram of the crosstalk between glucose signaling pathways in yeast. Yck I (Yck1 and Yck2) phosphorylates Mth1 and Std1 upon activation by glucose-bound Rgt2 and Snf3 glucose sensors. Phosphorylated Mth1 and Std1 are ubiquitinated by the SCFGrr1 complex and degraded by the proteasome. The PKA phosphorylation sites in the amino terminal region of Rgt1 are exposed and available for phosphorylation when Mth1 is degraded. Phosphorylated Rgt1 is dissociated form Ssn6-Tup1 and subsequently from DNA, leading to derepression of Rgt1 target genes, such as the HXT and HXK2 genes. The Rgt2/Snf3 pathway regulates itself through glucose-induction of STD1 gene expression. Consequently, the STD1 gene is expressed at the same time that the Std1 protein is degraded in response to glucose [13]. By contrast, glucose stimulates Mth1 degradation but also represses Mth1 expression via Mig1 and Mig2. Glucose uptake is required for the generation of the glucose repression signal that leads to inactivation of the Snf1 kinase [18]. Expression of the MIG2 gene is induced by glucose via the Rgt2/Snf3 pathway. Glucose-repression of SNF3 gene expression by Mig1 reflects the probable function of Snf3 as a high affinity glucose sensor, representing another important feature of the interaction between the glucose induction and repression pathways.
3.1, Proteasomal degradation of Mth1 via the Rgt2/Snf3 pathway
The glucose signal that leads to the proteasomal degradation of Mth1 and Std1 is generated by the two glucose transporter-like proteins Rgt2 and Snf3. These plasma membrane proteins are evolutionarily derived from glucose transporters, with 12 predicted transmembrane spanning domains, but appear to have lost the ability to transport glucose into the cell [71]. The supporting evidence is that growth defect of the hxt-null mutant deleted for the seven HXT genes (HXT1-7[23,26,72]) on glucose is suppressed by expression of any one of the seven HXT genes, but not of the RGT2 or SNF3 gene [73]. Furthermore, there are constitutive mutations in the glucose sensor genes (SNF3-1 and RGT2-1) that are thought to convert the proteins into the glucose-bound forms and cause glucose-independent expression of the HXT genes [73]. Thus, the prevailing view is that Rgt2 and Snf3 act as cell surface glucose receptors that initiate signal transduction in response to glucose [1,14].
Rgt2 and Snf3 are activated by different levels of glucose, presumably due to their different affinities for glucose. Rgt2 is activated by high levels of glucose and generates a signal that stimulates expression of low affinity glucose transporters such as Hxt1 and Hxt3. Snf3 generates a glucose signal in response to low levels of glucose, which stimulates expression of high affinity glucose transporters such as Hxt2 and Hxt4 [38]. A central player in the transduction of the signal generated by Rgt2 and Snf3 may be the yeast casein kinase I (Yck1 and Yck2), tethered to the plasma membrane through palmitoylation of the C-terminal Cys-Cys sequence by the palmitoyl transferase Akr1 [74-76]. A current view is that glucose-binding to the glucose sensors induces a change in their conformation that activates Yck1/2 [43], which in turn catalyzes phosphorylation of Mth1 and Std1 [43,45]. This phosphorylation marks Mth1 and Std1 for ubiquitination by the SCFGrr1 ubiquitin protein ligase and subsequent degradation by the proteasome [35,44,48]. This scenario is supported by the observation that glucose sensors interact with Mth1 and Std1 through their C-terminal, cytoplasmic tails [38,39]. However, a recent study demonstrated that Mth1 degradation occurs without a direct coupling between the glucose sensors and Yck1/2 [45]. Accordingly, there may be an unidentified mechanism for transduction of the glucose signal to Mth1 and Std1.
3.2, Repression of MTH1 gene expression by the Snf1-Mig1 pathway
Expression of the MTH1 gene is repressed by glucose via the Snf1-Mig1 pathway. The Snf1 kinase, the yeast homolog of mammalian AMP-activated protein kinase (AMPK), is a heterotrimer composed of the Snf1 catalytic subunit (α), the Snf4 regulatory subunit (γ), and one of the three β–subunits (Sip1, Sip2 or Gal83) that seem to serve as a scaffold in the Snf1 complex. Snf1 activation requires phosphorylation at Thr210 by its upstream kinases (Sak1, Tos3 and Elm1) when glucose is limited [77,78]. Glucose appears to stimulate dephosphorylation of Snf1 by the Reg1-Glc7 PP1 phosphatase at the same Thr210, leading to the conversion of Snf1 from an active to an inactive conformation [79,80]. Snf1 phosphorylates and negatively regulates the Mig1 repressor in glucose-limited conditions; glucose-induced inactivation of Snf1 leads to dephosphorylation and activation of Mig1. Activated Mig1 represses expression of many glucose-repressed genes, such as SUC2, GAL1 and GAL4[68,69]. Expression of the MTH1 gene is repressed by Mig1 and Mig2 in high glucose conditions [13]. Therefore, the extent and rate of Mth1 degradation is substantially reduced in cells missing Mig1 and Mig2 or lacking their binding site in the MTH1 promoter [48]. The purpose of this regulation is to ensure rapid removal of Mth1 from cells when glucose becomes available so as to enable prompt induction of HXT gene expression.
3.3, PKA-mediated Rgt1 phosphorylation disrupts its interaction with Ssn6-Tup1
The third pathway known to affect expression of HXT genes is the one that activates the cAMP-dependent protein kinase A (PKA) [81,82]. PKA is involved in many different cellular processes including cell growth, stress resistance, and metabolism [83-86]. PKA is inactive during non-fermentative growth, existing as a tetrameric holoenzyme composed of two catalytic subunits encoded by one of three redundant TPK genes (TPK1, TPK2 and TPK3) and two regulatory subunits encoded by BCY1[87,88]. Addition of glucose to yeast cells grown on a non-fermentable carbon source induces a rapid elevation of cAMP levels due to glucose activation of adenylate cyclase (Cyr1) via the Gpr1/Gpa2 and the Ras1/Ras2 pathways [89-92]. Binding of cAMP to the Bcy1 inhibitory subunit of PKA liberates the catalytic subunits, leading to their activation [93]. A role of PKA in regulation of Rgt1 is implicated from the observation that glucose-induction of HXT1 gene expression is abolished in cells with attenuated PKA activity [81,82]. PKA phosphorylates Rgt1 at four serine residues in the amino-terminal region of Rgt1 and modulates its function [81,94], resulting in dissociation of Rgt1 from Ssn6-Tup1 and eventually from the HXT promoters [67]. The observation that Rgt1 phosphorylation by PKA does not occur until the Rgt1-Mth1/Std1-Ssn6-Tup1 repressor complex is disrupted suggests that the PKA sites in Rgt1 are available for phosphorylation upon glucose-induced degradation of Mth1.
4. Crosstalk between glucose signaling pathways ensure stringent regulation of glucose utilization
The three glucose signaling pathways play distinctive but interacting roles for glucose induction of HXT gene expression (Fig. 3). Glucose induced inactivation of Mth1 is a crucial event for modulating Rgt1 function. Indeed, Mth1 levels are tightly controlled in a feed-forward manner by coordination of the glucose induction and repression pathways. In addition, the glucose repression signal that inactivates Snf1 kinase is generated through glucose metabolism [18].
4.1, Integration of glucose induction and glucose repression pathways in a regulatory network
A gene expression profiling study indicated that the Rgt2/Snf3 glucose induction and the Snf1-Mig1 glucose repression pathways are intertwined in a regulatory network [13]. Glucose stimulates the proteasome-mediated degradation of Mth1 via the Rgt2/Snf3 pathway while also reducing MTH1 expression via the Snf1-Mig1 pathway [35,43,48], reinforcing the inhibitory effect of glucose on Mth1 function and ensuring maximal glucose induction of Rgt1-repressed genes. Glucose inhibits Std1 function by stimulating its degradation; it also induces STD1 gene expression through the Rgt2/Snf3 pathway [13]. Therefore, the feedback control of STD1 gene expression acts to counteract the glucose-induced degradation of Std1 and thus provides for a rapid reestablishment of repression when glucose is depleted. This may contribute to the regulation of glucose repression, because Snf1 activity is enhanced by Std1 [95]. Mig2 is a glucose repressor and collaborates with Mig1 in repression of most glucose-repressed genes. Mig1 and Mig2 bind to the identical DNA sequences but are differently regulated [96,97]. Mig2, unlike Mig1, is not regulated by Snf1, but its expression is induced by glucose via the Rgt2/Snf3 pathway [13]. Therefore, glucose repression is a result of outputs from two glucose signal transduction pathways: the Mig1 component regulated by the Snf1 kinase and the Mig2 component regulated at the level of their transcription by the Snf3/Rgt2 signaling pathway. Expression of the SNF3 gene is repressed by the Snf1-Mig1 pathway in high levels of glucose, enabling Snf3 to function only in low glucose conditions [13]. Hxk2 is the most active hexokinase isoenzyme during growth on glucose in the cytoplasm, while it interacts with components repressing expression of several glucose-repressed genes in the nucleus [4]. Glucose induces expression of the HXK2 gene via the Rgt2/Snf3 pathway, providing another functional link between glucose induction and repression pathways [82].
4.2, The glucose repression signal is generated through glucose metabolism
Although the glucose-induced inactivation of Snf1 appears to be critical for establishing glucose repression, the nature of the glucose repression signal that inhibits Snf1 activity is not clearly defined. The AMP-activated protein kinase (AMPK) is known to be activated by an elevated AMP:ATP ratio upon glucose depletion [98]. The AMP:ATP ratio increases more than 200-fold upon glucose removal and decreases when glucose is added. However, Snf1 activity is not directly regulated by the AMP:ATP ratio [99]. Instead, it has been speculated that glucose-6-phosphate (G-6-P) might serve as a glucose repression signal based on the observations that: (1) Snf1 is constitutively active in a hxk1hxk2glk1 deletion mutant; (2) addition of the glucose analog 2-deoxyglucose, which can be phosphorylated but not further metabolized, to glycerol-grown cells results in the decreased activity of Snf1; (3) Snf1 activity is not affected by 6-deoxyglucose, which cannot be phosphorylated [100,101]. However, the observations that glucose repression may be related to the glucose concentration rather than glucose flux suggest that the glucose repression signal appears to be upstream of G-6-P [102,103]. In this regard, Ozcan has demonstrated that glucose repression of the Mig1 target genes is abolished in a mutant lacking all 17 hexose transporters or the Rgt2 and Snf3 glucose sensors [18]. Hence, regulation of glucose-induction of HXT gene expression plays an important role in the generation of the glucose repression signal, highlighting how glucose induction and glucose repression of gene expression is coordinated to ensure tight control of glucose uptake and metabolism.
5. Conclusions
Glycolysis is facilitated by increased glucose uptake. Glucose uptake in S. cerevisiae is a complex process, most notably due to proliferation of glucose transporters and multiple, parallel pathways for signaling glucose availability. In terms of displaying glycolytic phenotype, S. cerevisiae and tumor cells share several features. Expression of human glucose transporters (GLUTs) is upregulated by Hif-1α in tumor cells [104]. Hif1, like Rgt1, is regulated by ubiquitin-mediated degradation [105,106]. Snf1/AMPK is a low energy checkpoint, acting as the prime energy sensor in response to energy depletion, and its activation mechanism is conserved in eukaryotes. Several current diabetes therapeutics, such as metformin and thiazolidinediones (TZDs), are thought to lower blood glucose and increase insulin sensitivity by activating AMPK in peripheral tissues [107]. The upstream activating kinase of AMPK is LKB1, which is associated with Peutz-Jeghers cancer-susceptibility syndrome (PJS) [108]. More importantly, AMPK phosphorylates and activates the tumor suppressor TSC2, a critical negative regulator of mTOR (mammalian target of rapamycin) [109]. In addition, the finding that mTOR is a central activation point for expression of Hif-1α independent of oxygen levels implicates the role of AMPK in regulation of glucose transporters [110]. Thus, AMPK, like Snf1, is actively involved in glucose uptake and metabolism. These similarities indicate that further understanding of glucose sensing and signaling mechanisms in yeast may reveal the molecular basis of the enhanced glucose uptake in cancer cells and provide clues for developing therapeutic strategies.
Highlights.
The budding yeast S. cerevisiae, like tumor cells, exhibits aerobic glycolysis.
The yeast possesses a family of glucose transporters (HXTs) with different kinetic properties.
Expression of HXT genes regulated by the crosstalk between glucose signaling pathways
Acknowledgments
This work was supported by NIH Grant GM087470 from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rolland F, Winderickx J, Thevelein JM. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem Sci. 2001;26:310–317. doi: 10.1016/s0968-0004(01)01805-9. [DOI] [PubMed] [Google Scholar]
- 3.Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Busti S, Coccetti P, Alberghina L, Vanoni M. Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae. Sensors. 2010;10:6195–6240. doi: 10.3390/s100606195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol Metab. 2005;16:489–494. doi: 10.1016/j.tem.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Kolb H, Eizirik DL. Resistance to type 2 diabetes mellitus: a matter of hormesis? Nat Rev Endocrinol. 2012;8:183–192. doi: 10.1038/nrendo.2011.158. [DOI] [PubMed] [Google Scholar]
- 7.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 10.Mandelkern M, Raines J. Positron emission tomography in cancer research and treatment. Technol Cancer Res Treat. 2002;1:423–439. doi: 10.1177/153303460200100603. [DOI] [PubMed] [Google Scholar]
- 11.Crabtree HG. Observations on the carbohydrate metabolism of tumours. Biochem J. 1929;23:536–545. doi: 10.1042/bj0230536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagunas R. Energetic irrelevance of aerobiosis for S. cerevisiae growing on sugars. Mol Cell Biochem. 1979;27:139–146. doi: 10.1007/BF00215362. [DOI] [PubMed] [Google Scholar]
- 13.Kaniak A, Xue Z, Macool D, Kim JH, Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston M, Kim JH. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33:247–252. doi: 10.1042/BST0330247. [DOI] [PubMed] [Google Scholar]
- 15.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 16.Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozcan S. Two different signals regulate repression and induction of gene expression by glucose. J Biol Chem. 2002;277:46993–46997. doi: 10.1074/jbc.M208726200. [DOI] [PubMed] [Google Scholar]
- 19.Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192:73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolland F, Winderickx J, Thevelein JM. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002;2:183–201. doi: 10.1111/j.1567-1364.2002.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 21.Diaz-Ruiz R, Uribe-Carvajal S, Devin A, Rigoulet M. Tumor cell energy metabolism and its common features with yeast metabolism. Biochim Biophys Acta. 2009;1796:252–265. doi: 10.1016/j.bbcan.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Boles E, Hollenberg CP. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 23.Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J Biochem. 1997;245:324–333. doi: 10.1111/j.1432-1033.1997.00324.x. [DOI] [PubMed] [Google Scholar]
- 24.Gancedo JM. The early steps of glucose signalling in yeast. FEMS Microbiol Rev. 2008;32:673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- 25.Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol. 1995;16:157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x. [DOI] [PubMed] [Google Scholar]
- 27.Liang H, Gaber RF. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson BR, Boulton CA, Box WG, Graham NS, Lawrence SJ, Linforth RS, Smart KA. Carbohydrate utilization and the lager yeast transcriptome during brewery fermentation. Yeast. 2008;25:549–562. doi: 10.1002/yea.1609. [DOI] [PubMed] [Google Scholar]
- 29.Krampe S, Stamm O, Hollenberg CP, Boles E. Catabolite inactivation of the high-affinity hexose transporters Hxt6 and Hxt7 of Saccharomyces cerevisiae occurs in the vacuole after internalization by endocytosis. FEBS Lett. 1998;441:343–347. doi: 10.1016/s0014-5793(98)01583-x. [DOI] [PubMed] [Google Scholar]
- 30.Kim JH, Polish J, Johnston M. Specificity and regulation of DNA binding by the yeast glucose transporter gene repressor Rgt1. Mol Cell Biol. 2003;23:5208–5216. doi: 10.1128/MCB.23.15.5208-5216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH. Immobilized DNA-binding assay, an approach for in vitro DNA-binding assay. Anal Biochem. 2004;334:401–402. doi: 10.1016/j.ab.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 32.Kim JH. DNA-binding properties of the yeast Rgt1 repressor. Biochimie. 2009;91:300–303. doi: 10.1016/j.biochi.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panozzo C, Capuano V, Fillinger S, Felenbok B. The zinc binuclear cluster activator AlcR is able to bind to single sites but requires multiple repeated sites for synergistic activation of the alcA gene in Aspergillus nidulans. J Biol Chem. 1997;272:22859–22865. doi: 10.1074/jbc.272.36.22859. [DOI] [PubMed] [Google Scholar]
- 35.Flick KM, Spielewoy N, Kalashnikova TI, Guaderrama M, Zhu Q, Chang HC, Wittenberg C. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol Biol Cell. 2003;14:3230–3241. doi: 10.1091/mbc.E03-03-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosley AL, Lakshmanan J, Aryal BK, Ozcan S. Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J Biol Chem. 2003;278:10322–10327. doi: 10.1074/jbc.M212802200. [DOI] [PubMed] [Google Scholar]
- 37.Hubbard EJ, Jiang R, Carlson M. Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1972–1978. doi: 10.1128/mcb.14.3.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt MC, McCartney RR, Zhang X, Tillman TS, Solimeo H, Wolfl S, Almonte C, Watkins SC. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafuente MJ, Gancedo C, Jauniaux JC, Gancedo JM. Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol Microbiol. 2000;35:161–172. doi: 10.1046/j.1365-2958.2000.01688.x. [DOI] [PubMed] [Google Scholar]
- 40.Lakshmanan J, Mosley AL, Ozcan S. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr Genet. 2003;44:19–25. doi: 10.1007/s00294-003-0423-2. [DOI] [PubMed] [Google Scholar]
- 41.Tomas-Cobos L, Casadome L, Mas G, Sanz P, Posas F. Expression of the HXT1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signalling pathways. J Biol Chem. 2004;279:22010–22019. doi: 10.1074/jbc.M400609200. [DOI] [PubMed] [Google Scholar]
- 42.Polish JA, Kim JH, Johnston M. How the Rgt1 transcription factor of Saccharomyces cerevisiae is regulated by glucose. Genetics. 2005;169:583–594. doi: 10.1534/genetics.104.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moriya H, Johnston M. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc Natl Acad Sci U S A. 2004;101:1572–1577. doi: 10.1073/pnas.0305901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasula S, Jouandot D, 2, Kim JH. Biochemical evidence for glucose-independent induction of HXT expression in Saccharomyces cerevisiae. FEBS Lett. 2007;581:3230–3234. doi: 10.1016/j.febslet.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasula S, Chakraborty S, Choi JH, Kim JH. Role of casein kinase 1 in the glucose sensor-mediated signaling pathway in yeast. BMC Cell Biol. 11:17. doi: 10.1186/1471-2121-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamo FJ, Lafuente MJ, Gancedo C. The mutation DGT1-1 decreases glucose transport and alleviates carbon catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1994;176:7423–7429. doi: 10.1128/jb.176.24.7423-7429.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulte F, Wieczorke R, Hollenberg CP, Boles E. The HTR1 gene is a dominant negative mutant allele of MTH1 and blocks Snf3- and Rgt2-dependent glucose signaling in yeast. J Bacteriol. 2000;182:540–542. doi: 10.1128/jb.182.2.540-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JH, Brachet V, Moriya H, Johnston M. Integration of transcriptional and posttranslational regulation in a glucose signal transduction pathway in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:167–173. doi: 10.1128/EC.5.1.167-173.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabina J, Johnston M. Asymmetric signal transduction through paralogs that comprise a genetic switch for sugar sensing in Saccharomyces cerevisiae. J Biol Chem. 2009;284:29635–29643. doi: 10.1074/jbc.M109.032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varanasi US, Klis M, Mikesell PB, Trumbly RJ. The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol Cell Biol. 1996;16:6707–6714. doi: 10.1128/mcb.16.12.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz J, Marshall-Carlson L, Carlson M. The N-terminal TPR region is the functional domain of SSN6, a nuclear phosphoprotein of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4744–4756. doi: 10.1128/mcb.10.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith RL, Redd MJ, Johnson AD. The tetratricopeptide repeats of Ssn6 interact with the homeo domain of alpha 2. Genes Dev9. 1995:2903–2910. doi: 10.1101/gad.9.23.2903. [DOI] [PubMed] [Google Scholar]
- 53.Jabet C, Sprague ER, VanDemark AP, Wolberger C. Characterization of the N-terminal domain of the yeast transcriptional repressor Tup1. Proposal for an association model of the repressor complex Tup1 x Ssn6. J Biol Chem. 2000;275:9011–9018. doi: 10.1074/jbc.275.12.9011. [DOI] [PubMed] [Google Scholar]
- 54.Smith RL, Johnson AD. A sequence resembling a peroxisomal targeting sequence directs the interaction between the tetratricopeptide repeats of Ssn6 and the homeodomain of alpha 2. Proc Natl Acad Sci U S A. 2000;97:3901–3906. doi: 10.1073/pnas.070506797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sprague ER, Redd MJ, Johnson AD, Wolberger C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J19. 2000:3016–3027. doi: 10.1093/emboj/19.12.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith RL, Johnson AD. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- 57.Malave TM, Dent SY. Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol. 2006;84:437–443. doi: 10.1139/o06-073. [DOI] [PubMed] [Google Scholar]
- 58.Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev10. 1996:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 59.Davie JK, Edmondson DG, Coco CB, Dent SY. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem. 2003;278:50158–50162. doi: 10.1074/jbc.M309753200. [DOI] [PubMed] [Google Scholar]
- 60.Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 61.Mennella TA, Klinkenberg LG, Zitomer RS. Recruitment of Tup1-Ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot Cell. 2003;2:1288–1303. doi: 10.1128/EC.2.6.1288-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papamichos-Chronakis M, Gligoris T, Tzamarias D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 2004;5:368–372. doi: 10.1038/sj.embor.7400120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet. 2006;38:1446–1451. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desimone AM, Laney JD. Corepressor-directed preacetylation of histone H3 in promoter chromatin primes rapid transcriptional switching of cell-type-specific genes in yeast. Mol Cell Biol. 2010;30:3342–3356. doi: 10.1128/MCB.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hanlon SE, Rizzo JM, Tatomer DC, Lieb JD, Buck MJ. The stress response factors Yap6, Cin5, Phd1, and Skn7 direct targeting of the conserved co-repressor Tup1-Ssn6 in S. cerevisiae. PLoS ONE. 2011;6:e19060. doi: 10.1371/journal.pone.0019060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong KH, Struhl K. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 2011;25:2525–2539. doi: 10.1101/gad.179275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roy A, Shin YJ, Cho KH, Kim JH. Mth1 regulates the interaction between the Rgt1 repressor and the Ssn6-Tup1 corepressor complex by modulating PKA-dependent phosphorylation of Rgt1. Mol Biol Cell. 2013;24:1493–1503. doi: 10.1091/mbc.E13-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nehlin JO, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griggs DW, Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Treitel MA, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ozcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci U S A. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maier A, Volker B, Boles E, Fuhrmann GF. Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters. FEMS Yeast Res. 2002;2:539–550. doi: 10.1111/j.1567-1364.2002.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 73.Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Y, Davis NG. Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol Cell Biol. 2000;20:5350–5359. doi: 10.1128/mcb.20.14.5350-5359.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Babu P, Deschenes RJ, Robinson LC. Akr1p-dependent palmitoylation of Yck2p yeast casein kinase 1 is necessary and sufficient for plasma membrane targeting. J Biol Chem. 2004;279:27138–27147. doi: 10.1074/jbc.M403071200. [DOI] [PubMed] [Google Scholar]
- 77.Hong SP, Leiper FC, Woods A, Carling D, Carlson M. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc Natl Acad Sci U S A. 2003;100:8839–8843. doi: 10.1073/pnas.1533136100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nath N, McCartney RR, Schmidt MC. Yeast Pak1 kinase associates with and activates Snf1. Mol Cell Biol. 2003;23:3909–3917. doi: 10.1128/MCB.23.11.3909-3917.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 80.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JH, Johnston M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem. 2006;281:26144–26149. doi: 10.1074/jbc.M603636200. [DOI] [PubMed] [Google Scholar]
- 82.Palomino A, Herrero P, Moreno F. Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 2006;34:1427–1438. doi: 10.1093/nar/gkl028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Broach JR. RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 1991;7:28–33. doi: 10.1016/0168-9525(91)90018-l. [DOI] [PubMed] [Google Scholar]
- 84.Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 85.Rolland F, De Winde JH, Lemaire K, Boles E, Thevelein JM, Winderickx J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol Microbiol. 2000;38:348–358. doi: 10.1046/j.1365-2958.2000.02125.x. [DOI] [PubMed] [Google Scholar]
- 86.Thevelein JM, Gelade R, Holsbeeks I, Lagatie O, Popova Y, Rolland F, Stolz F, Van de Velde S, Van Dijck P, Vandormael P, Van Nuland A, Van Roey K, Van Zeebroeck G, Yan B. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem Soc Trans. 2005;33:253–256. doi: 10.1042/BST0330253. [DOI] [PubMed] [Google Scholar]
- 87.Cannon JF, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1987;7:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 89.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 90.Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, Matsumoto K, Kaziro Y, Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60:803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- 91.Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, Nauwelaers D, de Winde JH, Gorwa MF, Colavizza D, Thevelein JM. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Versele M, de Winde JH, Thevelein JM. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Edelman AM, Blumenthal DK, Krebs EG. Protein serine/threonine kinases. Annu Rev Biochem. 1987;56:567–613. doi: 10.1146/annurev.bi.56.070187.003031. [DOI] [PubMed] [Google Scholar]
- 94.Jouandot D, 2, Roy A, Kim JH. Functional dissection of the glucose signaling pathways that regulate the yeast glucose transporter gene (HXT) repressor Rgt1. J Cell Biochem. 2011;112:3268–3275. doi: 10.1002/jcb.23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuchin S, Vyas VK, Kanter E, Hong SP, Carlson M. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics. 2003;163:507–514. doi: 10.1093/genetics/163.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lutfiyya LL, Iyer VR, DeRisi J, DeVit MJ, Brown PO, Johnston M. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–1391. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lutfiyya LL, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 99.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 100.Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 2001;15:1104–1114. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hedbacker K, Carlson M. Regulation of the nucleocytoplasmic distribution of Snf1-Gal83 protein kinase. Eukaryot Cell. 2006;5:1950–1956. doi: 10.1128/EC.00256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meijer MM, Boonstra J, Verkleij AJ, Verrips CT. Glucose repression in Saccharomyces cerevisiae is related to the glucose concentration rather than the glucose flux. J Biol Chem. 1998;273:24102–24107. doi: 10.1074/jbc.273.37.24102. [DOI] [PubMed] [Google Scholar]
- 103.Ye L, Kruckeberg AL, Berden JA, van Dam K. Growth and glucose repression are controlled by glucose transport in Saccharomyces cerevisiae cells containing only one glucose transporter. J Bacteriol. 1999;181:4673–4675. doi: 10.1128/jb.181.15.4673-4675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem. 1995;270:29083–29089. doi: 10.1074/jbc.270.49.29083. [DOI] [PubMed] [Google Scholar]
- 105.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 107.Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 109.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 110.Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]


