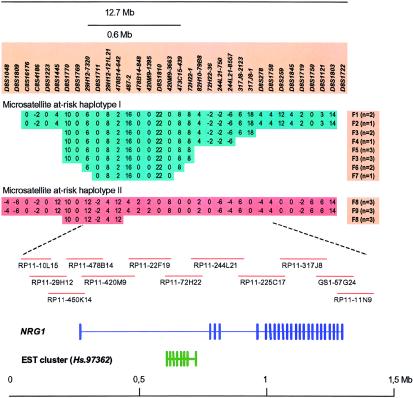
Figure 3.
Microsatellite at-risk haplotypes on chromosome 8p and known genes within a region showing haplotype sharing between 9 of the 33 linkage families. Extensive sharing of two microsatellite haplotypes between patients from the linkage families is shown at the top. Haplotypes were reconstructed by application of the Allegro program (Gudbjartsson et al. 2000), using available family members to derive the phase. Key markers in the haplotypes are shown, and the size of the region is indicated. Families carrying the haplotypes are labeled F1–F9, and the number of affected individuals in each family carrying that haplotype is given in parentheses. Maximum haplotype sharing between families is 9.5 Mb for haplotype I and 11.4 Mb for haplotype II. Shared haplotypes between families narrow the region of interest to 600 kb between markers 29H12-7320 and 473C15-439, indicated by a bar (microsatellite haplotype I). The location of a BAC contig covering 1.5 Mb of the locus region is indicated. The sequence of GS1-57G24 was obtained from the public domain (GenBank accession number AF128834), but we sequenced the other BACs shown. The positions of NRG1 and an EST cluster of unknown function (Hs.97362) are schematically shown in relation to the BACs. Exons are indicated by vertical bars.