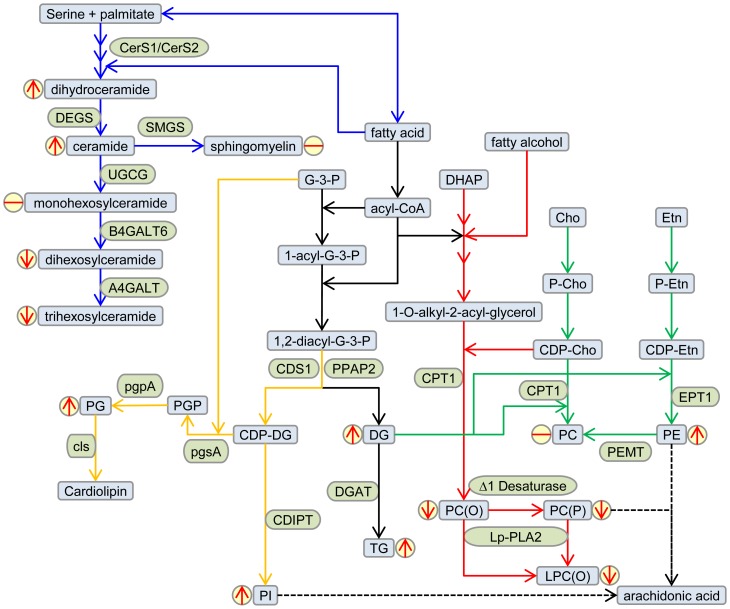
Figure 5. Metabolic pathways altered in type 2 diabetes.
Partial lipid metabolic pathways show the major lipids associated with T2D (blue boxes) and the enzymes involved (green boxes). The sphingolipid (blue arrows), cardiolipin (orange arrows), triacylglycerol (black arrows), plasmalogen (red arrows) and phosphatidylcholine/phosphatidylethanolamine (green arrows) biosynthetic pathways are shown. The direction of association between lipids and T2D is indicated by the red arrows in yellow circles. Only partial pathways have been shown for clarity. Metabolite abbreviations: Cho, choline; DG, diacylglycerol; DHAP, dihydroxyacetonephosphate; Etn, ethanolamine; LPC(O), lysoalkylphosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PC(O), alkylphosphatidylcholine; PC(P), alenylphosphatidylcholine; PG, phosphatidylglycerol; PGP, phosphatidylglycerolphosphate; PI, phosphatidylinositol; TG, triacylglycerol. Enzyme abbreviations: A4GALT, lactosylceramide 4-alpha-galactosyltransferase; B4GALT6, beta-1,4-galactosyltransferase 6; CDIPT, CDP-diacylglycerol-inositol 3-phosphatidyltransferase; CDS1, phosphatidate cytidylyltransferase; CerS, ceramide synthasecls, cardiolipin synthase; CPT1, diacylglycerol cholinephosphotransferase; DEGS, sphingolipid delta-4 desaturase; DGAT, diacylglycerol O-acyltransferase; EPT1, PEMT, phosphatidylethanolamine N-methyltransferase; ethanolaminephosphotransferase, pgpA, phosphatidylglycerophosphatase A; pgsA, CDP-diacylglycerol – glycerol-3-phosphate 3-phosphatidyltransferase; PPAP2, phosphatidate phosphatase; SMGS, sphingomyelin synthase; UGCG, ceramide glucosyltransferase.