Figure 4. Process of open reading frame (ORF) selection using the AGM13 helper phage.
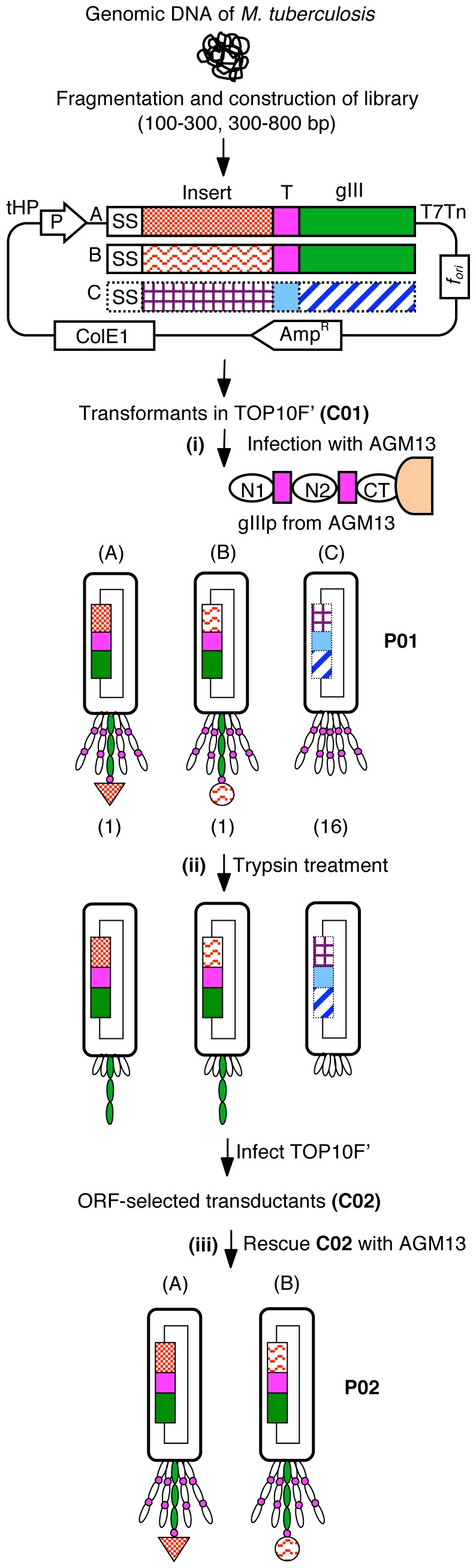
The gene-fragment library is constructed in a phagemid vector by inserting the fragments between the PelB signal sequence and full length native gIIIP, and transformants (C01, primary unselected library) are obtained in E. coli TOP10F’. (i) The C01 cells are used for producing phages using AGM13, which has a trypsin cleavage site (T) in the linker regions between the N1 and N2, and N2 and CT domains of gIIIP. During rescue, three types of phages are produced: A, Phages displaying protein matching with the M. tuberculosis proteome (genic clones) expressed as fusion protein with full length trypsin-resistant gIIIP; B, phages displaying protein not matching with the M. tuberculosis proteome (non-genic clone) but expressed as fusion protein with full length trypsin-resistant gIIIP. C, phages not displaying any protein as the insert is out-of-frame with the signal sequence and trypsin-resistant gIIIP, which carry only AGM13-encoded trypsin-sensitive gIIIP. (ii) Trypsin treatment of the phage population renders non-displaying phages (type C) non-infectious, while A and B become fully infectious due to removal of the displayed protein by cleavage of the trypsin site present between gIIIP and the fusion partner. Subsequent infection of the trypsin-treated library produces ORF-selected transductants (C02). (iii) Upon rescue with AGM13 or any other helper phage, the C02 transductants produce ORF-selected phages displaying protein, which could be from genic or non-genic DNA sequences.
