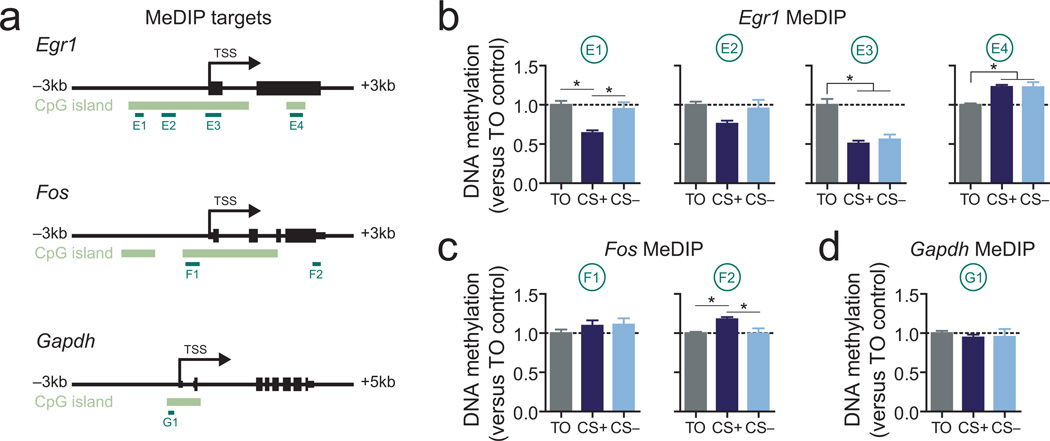
Figure 3.
Reward-related memory formation alters VTA DNA methylation profiles at Egr1 and Fosa, Gene targets for MeDIP assay. Annotated lines below genes illustrate loci for gDNA primer pairs. b-d, MeDIP reveals site-specific learning and reward-related changes in DNA methylation at Egr1 and Fos (n = 8 per group; one-way ANOVA: main effect of training for E1, E3, E4, F2 sites, P < 0.008 for each comparison; Tukey post-hoc tests revealed significant group differences (*P < 0.05)). Error bars represent s.e.m.