Fig. 3.
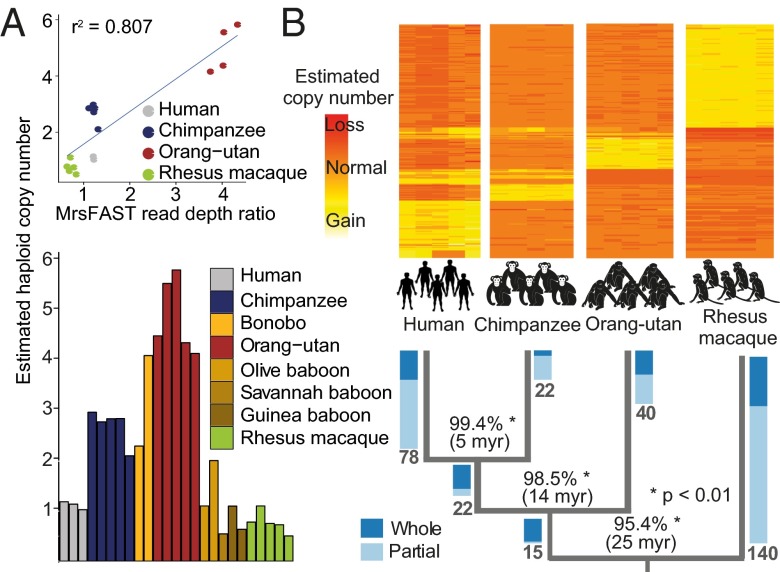
Investigation of fixed gene duplications. (A) qPCR verification of SUZ12 gene duplication. (Upper) Correlation of micro-read substitution-only Fast Alignment Search Tool (mrsFast) read-depth ratios (x axis) and estimated haploid copy numbers by qPCR (y axis). (Lower) qPCR results in an extended panel with eight different primate species. (B) Timing of the occurrence of fixed gene duplications in primate evolution. The heat map depicts mrsFAST read-depth ratios of fixed gene duplications that were timed. Rows represent timed orthologous genes, and columns represent individual samples. Yellow colors indicate higher read-depth ratios (>1) corresponding to a gain. Orange colors indicate read-depth ratios of ~1 corresponding to two diploid copies. Red colors indicate read-depth ratios < 1 corresponding to an inferred loss. In Lower, numbers in bold at tree edges represent the numbers of genes timed for a specific tree branch. Blue-colored bars represent ratios of timed whole- and partial gene duplications on a specific branch. Percentages at tree branches depict the mean sequence identity between duplicated paralogs on each of the branches (computed based on segmental duplication overlap).