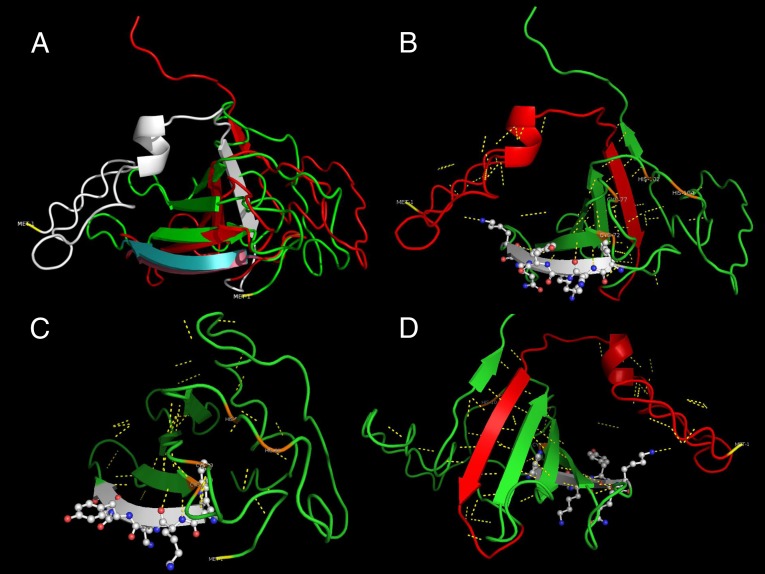
Fig. 5.
3D structure comparison and functional prediction of SbWRKY and SpWRKY peptides. (A) Superimposition of SbWRKY (green) and SpWRKY (red) transcription factors aligned at the first four amino acid residues of WRKY domains. Structure of the SbWRKY domain (cyan) departs very rapidly from that of SpWRKY domain (purple). The truncated 44 amino acid residues of SpWRKY are shown in white. (B) Conserved WRKY DNA binding domain is shown on second β-plated sheet (white). The first β-sheet in the truncated section in SbWRKY (red) intercalates between the fifth and sixth β-sheets forming a ribcage supported by polar interactions (yellow interrupted lines). The model also predicts a zinc-finger domain among Cys-72, Cys-77, His-58, and His-104 residues (orange). (C) Model of the truncated SbWRKY. Lack of the first β-sheet disrupts the integrity. The predicted zinc-finger domain maintains it functional structure among the four residues (orange). (D) Closer view of the β-sheets after 180° rotation of the SpWRKY model. There are 17 polar interactions, of which 15 maintain β-sheet orientation. The remaining two interactions are with the WRKY domain and might play a central role in DNA binding.