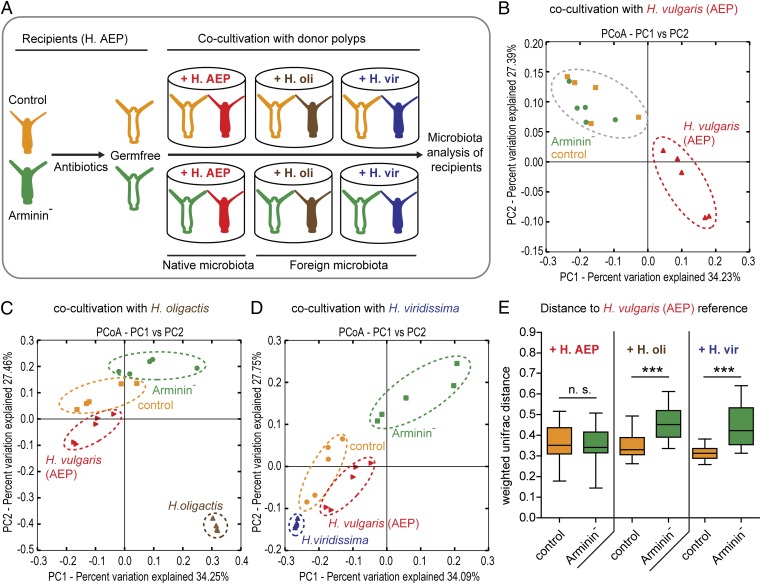
Fig. 5.
Species-specific arminins select for species-specific bacteria. (A) Schematic representation of the experimental design. Germ-free control polyps as well as germ-free Arminin− polyps [= transgenic H. vulgaris (AEP)] were cocultivated with either H. vulgaris (AEP), H. oligactis (H.oli), or H. viridissima (H.vir) polyps for 5 wk. (B–D) Bacterial communities were clustered using PCoA of the weighted Unifrac distance matrix. All three PCoA plots are based on the same distance matrix containing all 45 samples, and therefore all samples are presented relative to each other. The percent variation explained by the principle coordinates is indicated at the axes. (B) When inoculated with native bacteria retrieved from H. vulgaris (AEP), control and Arminin− polyps show no differences in bacterial recolonization. (C and D) When cocultivated with H. oligactis (C) or H. viridissima (D), control and Arminin− polyps cluster separately, with recolonized control polyps clustering proximate to H. vulgaris (AEP) reference polyps. (E) Comparison of the weighted Unifrac distances to the native H. vulgaris (AEP) microbiota. Control polyps infected with a H. oligactis or H. viridissima microbiota show significantly lower Unifrac distances to their bona fide wild-type status than Arminin− polyps. Statistics were carried out using two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001.