Fig. 2.
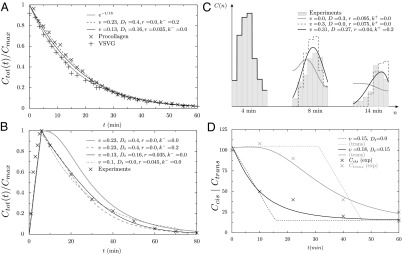
Quantitative analysis of data from different experimental protocols using a numerical solution of Eq. 3. (A and B) Optical microscopy assays. A whole Golgi FRAP experiment probing the exit of tagged proteins from the Golgi following (A) a steady influx, abruptly stopped at 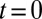 , of a small transmembrane protein (VSVG) and a large soluble protein aggregate (procollagen), and (B) a short influx, starting at
, of a small transmembrane protein (VSVG) and a large soluble protein aggregate (procollagen), and (B) a short influx, starting at 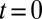 and stopping at
and stopping at 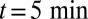 , of VSVG (10).
, of VSVG (10).  was set to zero in the fits because it does not influence the early relaxation. (C and D) EM assays. (C) Pulse-chase experiment for VSVG (13). Setting either convection (gray curve) or diffusion (dashed curve) to zero cannot reproduce the data. Fits are constrained so that the total protein concentration matches the data at
was set to zero in the fits because it does not influence the early relaxation. (C and D) EM assays. (C) Pulse-chase experiment for VSVG (13). Setting either convection (gray curve) or diffusion (dashed curve) to zero cannot reproduce the data. Fits are constrained so that the total protein concentration matches the data at 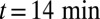 .
. 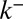 was set to zero because it has the same effect as r. (D) Evolution of the concentration of procollagen aggregates in the cis (black) and trans (gray) face of the Golgi upon sudden blockage of ER secretion (exiting wave experiment) (9). Data are in percentage of the concentration in normal conditions (steady ER secretion), and are not sensitive to exit rate. More information on the fitting procedure and experimental uncertainty is given in SI Appendix.
was set to zero because it has the same effect as r. (D) Evolution of the concentration of procollagen aggregates in the cis (black) and trans (gray) face of the Golgi upon sudden blockage of ER secretion (exiting wave experiment) (9). Data are in percentage of the concentration in normal conditions (steady ER secretion), and are not sensitive to exit rate. More information on the fitting procedure and experimental uncertainty is given in SI Appendix.
