Abstract
Pathogenic uncultivable treponemes, similar to syphilis-causing Treponema pallidum subspecies pallidum, include T. pallidum ssp. pertenue, T. pallidum ssp. endemicum and Treponema carateum, which cause yaws, bejel and pinta, respectively. Genetic analyses of these pathogens revealed striking similarity among these bacteria and also a high degree of similarity to the rabbit pathogen, T. paraluiscuniculi, a treponeme not infectious to humans. Genome comparisons between pallidum and non-pallidum treponemes revealed genes with potential involvement in human infectivity, whereas comparisons between pallidum and pertenue treponemes identified genes possibly involved in the high invasivity of syphilis treponemes. Genetic variability within syphilis strains is considered as the basis of syphilis molecular epidemiology with potential to detect more virulent strains, whereas genetic variability within a single strain is related to its ability to elude the immune system of the host. Genome analyses also shed light on treponemal evolution and on chromosomal targets for molecular diagnostics of treponemal infections.
Keywords: Treponema pallidum, T pallidum ssp. Pertenue, T. pallidum ssp. endemicum, T. paraluiscuniculi, whole genome sequencing, molecular evolution, molecular diagnostics
1. Introduction
The genus Treponema (Krieg et al., 2010) comprises several human pathogens that cause chronic infections including several species of oral treponemes (e.g. T. denticola, T. parvum, and T. putidum;Visser and Ellen, 2011). Another member of the genus Treponema, Treponema pallidum subspecies pallidum (TPA) is the causative agent of the sexually transmitted disease syphilis, a chronic, multi-stage, human infection characterized by variable clinical symptoms. Compared to treponemes isolated from the mouth (subgingival plaque), not a single strain of T. pallidum has been steadily propagated under in vitro conditions. Moreover, genome sequencing has revealed considerably larger genomes of oral treponemes (e.g. 2.84 Mb for T. denticola, (MacDougall and Girons, 1995; Seshadri et al., 2004) compared to T. pallidum strains (1.14 Mb; Fraser et al., 1998). In this review, we will focus on the genetic diversity of obligatory pathogenic Treponema pallidum strains and on related uncultivable treponemes.
Pathogenic treponemes, similar to syphilis-causing TPA, include Treponema pallidum subspecies pertenue (TPE), the causative agent of yaws, Treponema pallidum subspecies endemicum (TEN), the causative agent of endemic syphilis, and Treponema carateum, the causative agent of pinta. Besides these human pathogens, a rabbit pathogen, Treponema paraluiscuniculi, has been shown to be very similar to syphilis treponeme (Strouhal et al., 2007). Pathogenic treponemes of this group differ in two principal etiopathogenic parameters including host range and degree of invasivity; with TPA being the most invasive, TPE being moderately invasive and T. carateum being non-invasive. Treponema paraluiscuniculi is not pathogenic to humans; whereas TPA and TPE strains can infect both humans and rabbits (isolates of T. carateum have not yet been described).
Sequencing of the whole TPA Nichols genome in 1998 (Fraser et al., 1998), started an era of whole genome analyses of pathogenic treponemes. Results, to date, have increased our understanding of pathogenic treponemes and provided possibilities for molecular diagnostics of syphilis and yaws. Moreover, whole genome sequences of uncultivable pathogens have been used in a number subsequent studies (Brinkmann et al., 2006, Brinkmann et al., 2008; McGill et al., 2010; McKevitt et al., 2003, 2005; Mikalová et al., 2010; Šmajs et al., 2002, 2005; Strouhal et al., 2007; Titz et al., 2008; Weinstock et al., 2000a).
2. Low level of genetic diversity in noncultivable pathogenic treponemes
In many pathogenic bacteria, including both Gram-negative and Gram-positive bacteria, pathogenicity islands are important bacterial virulence determinants encoding a number of virulence factors (Schmidt and Hensel, 2004). Genes present in the genomic islands can be transferred from bacterium to bacterium via horizontal gene flux and the foreign DNA can also be incorporated into the host DNA. In addition, other mobile genetic elements, including conjugative and other plasmids, bacteriophages, transposons, and insertion elements can encode virulence factors. In contrast to this strategy, several pathogenic bacteria appear not to contain pathogenicity islands, including Mycobacterium spp., Chlamydia spp., most streptococcal species, and spirochetes. The common theme of pathogenic bacteria lacking pathogenicity islands is adaptation to a specific host environment, usually accompanied with genome size reduction and loss of the ability to replicate outside a host. Adaptation to a single or only a few hosts is usually accompanied by reduced horizontal gene transfer. Moreover, several bacterial pathogens have a highly clonal population structure (monomorphic bacterial pathogens), including Bacillus anthracis (Van Ert et al., 2007), Burkholderia mallei (Godoy et al., 2003), Chlamydophila pneumoniae (Rattei et al., 2007), Mycobacterium leprae (Monot et al., 2005), Mycobacterium tuberculosis complex (DosVultos et al., 2008), Salmonella enterica serovar Typhi (Kidgell et al., 2002; Roumagnac et al., 2006) and Yersinia pestis (Achtman et al., 1999, 2004). Many of these monomorphic bacteria have unknown immediate ancestors (Achtman, 2008). One of the potential explanations for such genetic monomorphism is the fact that humans are often secondary hosts for monomorphic pathogenic bacteria and therefore represent a rather recent branch off the main evolutionary course of these bacteria (Achtman, 2008). The genome size of pathogenic treponemes is much reduced, being approximately one fourth to one fifth the size of the E. coli genome. Pathogenic uncultivable treponemes can be classified as genetically monomorphic bacteria (Achtman, 2008) in the process of adaptation to a narrow host environment without known mechanisms of horizontal gene transfer and no obvious ancestors. Relatively subtle genetic changes are expected to determine the differences in treponemal host specificity and pathogenicity (see Table 1).
Table 1.
Genetic identity among treponemal strains compared to the Nichols strain based on whole genome sequencing (WGS) or whole genome fingerprinting (WGF).
| Strain | Place and year of isolation [reference] | Genome size as revealed by WGS (nt) | Estimated genome size by WGF (kb) | Reference (GenBank acc. no.) | Estimated genome sequence identity with Nichols (%)* |
|---|---|---|---|---|---|
| TPA Nichols | Washington, DC; 1912 Nichols and Hough, 1913 | 1,138,011 1,139,631** | Fraser et al., 1998 (AE000520.1) | 100 | |
| TPA Chicago | Chicago?; 1951 Turner and Hollander, 1957 | 1,139,281 | Giacani et al., 2010 (CP001752.1) | 99.95 | |
| TPA DAL-1 | Dallas; 1991 Wendel et al., 1991 | 1139.9 | Mikalová et al., 2010 | 99.97 | |
| TPA SS14 | Atlanta; 1977 Stamm et al., 1983 | 1,139,457 | Matějková et al., 2008 (CP000805.1) | 99.89 | |
| TPA Mexico A | Mexico; 1953 Turner and Hollander, 1957 | 1140.0 | Mikalová et al., 2010 | 99.88 | |
| TPE Samoa D | Western Samoa; 1953 Turner and Hollander, 1957 | 1,139,330 | D. Čejková et al., 2011 (CP002374) | 99.65 | |
| TPE CDC-2 | Akorabo, Ghana; 1980 Liska et al., 1982 | 1,139,744 | D. Čejková et al., 2011 (CP002375) | 99.64 | |
| TPE Gauthier | Congo; 1960 Gastinel et al., 1963 | 1,139,441 | D. Čejková et al., 2011 (CP002376) | 99.67 | |
| Fribourg-Blanc | Guinea; 1966 Fribourg-Blanc and Mollaret, 1969 | 1140.4 | Mikalová et al., 2010 | 99.57*** | |
| T. paraluiscuniculi Cuniculi A | Baltimore; ? Jacobsthal, 1920 | 1,133,390 | Šmajs et al., 2011 (CP002103) | 98.10 |
Genome sequence identity was calculated from whole genome sequence alignments including gaps in alignments.
The published genome sequence (Fraser et al., 1998) was modified by addition of insertion of a tprK-like sequence (1.3 kb) in the intergenic region between TP0126 and TP0127 (Šmajs et al., 2002) and addition 7 repetitive sequence (60 bp) in the arp gene (TP0433-TP0434).
identity based on whole genome fingerprinting (Mikalová et al., 2010)
Pathogenic uncultivable treponemes were originally considered as separate species based on disease symptomatology and epidemiology. Moreover, these bacteria clearly differed from nonpathogenic T. phagedenis and T. refringens treponemes (Miao and Fieldsteel, 1978). Later, it became clear that TPA and TPE strains were almost identical in DNA hybridization experiments (Miao and Fieldsteel, 1980) and this fact led to reclassification of both organisms into a single species (Smibert, 1984). Recent data from the T. paraluiscuniculi genome analysis (Šmajs et al., 2011) revealed that the genetic diversity between this strain and TPA strains, on a genome wide scale, was less than 2% and thus T. paraluiscuniculi represents a T. pallidum subspecies, rather than a new species. This fact further supports the genetic compactness of uncultivable treponemal pathogens and indicates that small genetic changes can result in profound changes in pathogenesis and host range. Therefore, every nucleotide change should be considered to have the potential for changing bacterial virulence.
3. Genetic differences between human T. pallidum pathogens and the rabbit pathogen T. paraluiscuniculi
Spirochetes that cause rabbit genital lesions were first denoted as Spirochaeta paralues-cuniculi (syphilis-like spirochete; Jacobsthal, 1920). Since that time, several papers have described rabbit infections with this organism, occurring both in the wild, as well as among laboratory animals; the pathogen has since been renamed T. paraluiscuniculi (DiGiacomo et al., 1983, 1984, 1985; Smith and Pesetsky, 1967). Rabbit venereal spirochetosis can be sexually transmitted and leads to mucocutaneous lesions of the genitoanal region, which are characterized by erythema, edema and/or crusting ulcers. In fact, T. paraluiscuniculi appears to be well adapted to rabbits, which develop active infections resembling syphilis in humans. However, unlike syphilis, no vertical transfer of T. paraluiscuniculi in rabbits has been documented. T. paraluiscuniculi is not infectious to humans and the non-infectivity was experimentally demonstrated via intradermal inoculation of three volunteers (Graves and Downes, 1981; Levaditi et al., 1921). In contrast to humans, intradermal inoculation of rabbits with T. paraluiscuniculi leads to chronic lesions with a serological response to T. paraluiscuniculi
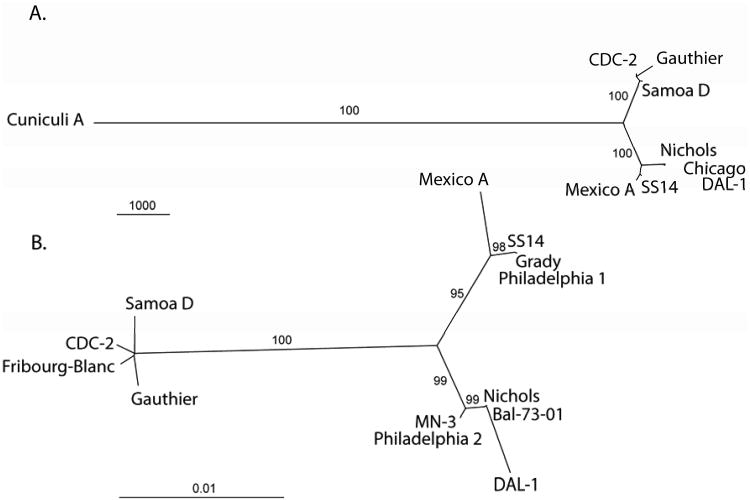
Previous studies indicated that the 5′ and 3′ flanking regions of the 15-kDa lipoprotein gene (tpp15) distinguished the human pathogens from T. paraluiscuniculi strains (Centurion-Lara et al., 1998), thus providing the first evidence of genetic differences between these pathogens. Other studies (Giacani et al., 2004; Grey et al., 2006) identified heterogeneity in the paralogous tpr genes. Whole genome fingerprinting and microarray DNA analysis of the T. paraluiscuniculi Cuniculi A strain revealed indels and prominent sequence changes in 38 gene homologs and six intergenic regions of the Cuniculi A genome compared to the genome of T. pallidum subsp. pallidum Nichols (Strouhal et al., 2007). Interestingly, most of the observed differences were identified in tpr genes or in their vicinity. In addition to tpr genes, sequence analysis of heterologous chromosomal regions identified 14 additional genes with frameshift mutations (Strouhal et al., 2007). Harper et al. (2008a) identified over 50 nucleotide changes between TPA strains and T. paraluiscuniculi; additionally, differences were also observed in the arp gene (Harper et al., 2008b). The genome sequence of Treponema paraluiscuniculi, strain Cuniculi A, was recently determined (Šmajs et al., 2011) using a combination of several high-throughput sequencing methods. An unrooted tree constructed from whole genome sequence alignments is shown in Fig. 1A. The T. paraluiscuniculi genome was found to contain a high number of pseudogenes and gene fragments (51) and the genome size was reduced compared to the TPA and TPE genomes. The Cuniculi A genome (1,133,390 bp) is 4.6, 5.9, and 6.1 kb smaller than the genome sizes of TPA Nichols (1,138,006 bp) (Fraser et al., 1998), TPA Chicago (1,139,281 bp; Giacani et al., 2010), and TPA SS14 (1,139,457 bp; Matějková et al., 2008), respectively. Since it is known that at least part of the Nichols population contains a 1.3 kb tprK-like insertion, localized in the intergenic region between TP0126 and TP0127 gene (Šmajs et al., 2002), and that the arp gene (TP0433-TP0434) contains 14 repetitive sequence (Pillay et al., 1998) motifs (60 bp in length) instead of the 7 repetitions reported by Fraser et al. (1998), the Cuniculi A genome is, in fact, 6.2 kb smaller than the Nichols genome. Compared to TPA Nichols, Chicago, and the SS14 genomes, whole genome nucleotide diversity between these genomes and the Cuniculi A genome ( ± SD) ranged from 0.01016 ± 0.00508 to 0.01028 ± 0.00514, indicating an extremely close relatedness between the human non-pathogenic strain and syphilis strains.
Fig. 1.
A. An unrooted tree constructed from whole genome sequence alignments using the maximum parsimony method. The bar scale corresponds to 1000 nt changes. Bootstrap values based on 1,000 replications are shown next to branches. The tree is very similar to that obtained following analysis of binary restriction target site data (Mikalová et al., 2010).
B. An unrooted tree constructed from concatenated nucleotide sequences of TP0136, TP0326, TP0488, and TP0548 genes using the maximum likelihood method. The length of sequenced regions ranged from 8342 to 8412 nucleotides. The bar scale corresponds to 0.01 nt changes per site. Bootstrap values based on 1,000 replications are shown next to branches. The TPA strains cluster into two separate groups, one containing the Nichols and the second the SS14 strain. In addition to strains listed in the Table 1, TPA strains Grady, Philadelphia 1, Philadelphia 2, MN-3, and Bal-73-01 are shown.
Sequencing of T. paraluiscuniculi genome (Šmajs et al., 2011) allowed identification of groups of treponemal genes, important in TPA strains for infection of human hosts. Two features stand out; first, 134 (13.2%) of Cuniculi A genes encoded identical proteins compared to Nichols proteins, indicating negative selection of corresponding genes and conservation of proteins. These proteins were mostly involved in translation and general metabolism; however, for 35 of them, no function was predicted, suggesting as yet unknown essential functions in treponemal metabolism. Second, 84 Cuniculi A genes (32 with predicted function) were found to contain frameshifts or major deletions or other major sequence changes (defined as changes causing continuous amino acid replacements comprising 10 and more residues or 20 and more dispersed amino acid replacements). When counting the number of affected genes in different functional groups and comparing them to the number of affected genes linked to the general metabolism group, 3 groups of genes including (i) virulence factors, (ii) genes with an unknown function and (iii) genes involved in DNA metabolism, contained significantly more frameshifts and major sequence changes. Since the median transcript levels, during experimental TPA Nichols rabbit infection (Šmajs et al., 2005) of affected genes with unknown function, was considerably higher than the median gene expression rate of all genes of unknown function (1.46 versus 0.86), these genes represent promising candidates for important virulence factors of T. pallidum Moreover, affected genes linked to DNA metabolism could suggest their possible role in the acceleration of T. paraluiscuniculi evolution.
4. Genetic differences between human T. pallidum subspecies pallidum and T. pallidum subspecies pertenue pathogens
Treponema pallidum ssp. pertenue (TPE) is the causative agent of yaws, a tropical disease with an estimated prevalence of about 2 million cases worldwide (World Health Organization, 1998). TPE was discovered in 1905 (Castellani, 1905) and unlike syphilis, yaws is not vertically transmitted and is mostly characterized by skin, joint, soft tissue and bone affections. In general, yaws treponemes are considered less virulent compared to syphilis treponemes and this fact is likely encoded in the TPE genome.
DNA hybridizations performed with TPA and TPE strains, in 1980, revealed that TPE strain Gauthier and TPA strain Nichols were identical within the limits of the resolution of the technique (i.e. about 2% of the genome differences; Miao and Fieldsteel, 1980). This observation resulted in reclassification of both organisms into a single species (Smibert, 1984).
The first papers describing genetic differences between syphilis and yaws treponemes appeared about 20 years ago (Noordhoek et al., 1989, 1990). However, the observed difference in gene TP1038 (tpf-1 gene in TPA, tyf-1 gene in TPE) at position 122, between TPA Nichols and TPE CDC 2575, was not specific for TPE strains. Walker et al. (1995) cited unpublished data indicating a difference in the 16S rRNA gene between a single TPA and a single TPE strain. Later, Centurion-Lara et al. (1998) and Stamm et al. (1998) described genetic changes differentiating TPA and TPE strains in the 5′ and 3′ flanking regions of tpp15 and in the tprJ gene, respectively. Differences between TPA and TPE strains were also found in the gpd gene (TP0257) at position 579, by Cameron et al. (1999), in tp92 (Cameron et al, 2000), and in tprI and tprC genes (Centurion-Lara et al., 2006).
Harper et al. (2008a) found over a dozen nucleotide changes differentiating TPA and TPE strains and also TPA and TEN strains. Moreover, genetic analysis of the arp gene revealed repeat motifs differentiating venereal and non-venereal strains (Harper et al., 2008b).
The work of Mikalová et al. (2010) compared genomes of 4 TPA treponemes (Nichols, SS14, DAL-1 and Mexico A) to 3 TPE strains (Samoa D, CDC-2 and Gauthier), and the Fribourg-Blanc isolate (Fribourg-Blanc and Mollaret, 1969) using the whole genome fingerprinting technique (WGF; Strouhal et al., 2007; Weinstock et al., 2000a). Restriction target site analysis comprising detection of 1773 individual restriction sites, found a similar structure in all investigated genomes. The unclassified simian Fribourg-Blanc was found to cluster with TPE strains but not with TPA strains. Most of the identified genetic differences between TPA and TPE strains were localized in six chromosomal regions, mostly around tpr genes. Based on WGF data, the estimated genome sequence identity between TPA and TPE strains were considered to be 99.63% or higher.
Complete genome sequences of three TPE strains (Samoa D, CDC-2, and Gauthier) were determined using next-generation sequencing techniques (Čejková et al., 2011). The genome lengths ranged between 1,139,330 bp and 1,139,744 bp and a similar genome structure was found among TPE strains as well as between TPA and TPE strains. The overall identity between TPA and TPE genomes was found to be over 99.64% (Table 1). Compared to four TPA strains (Nichols, DAL-1, SS14, and Chicago), changes consistently present in all three TPE strains were considered potentially responsible for the increased virulence of syphilis strains. Sequencing of 3 pertenue genomes (Čejková et al., 2011) revealed 692 out of 983 TPE protein-coding genes (70.4%) to encode identical proteins or identical proteins with strain specific changes, 194 (19.7%) genes to encode proteins with 1 amino acid substitution, and 97 (9.9%) TPE genes to encode proteins containing two or more amino acid replacements and/or other major sequence changes. Similar to results found in the Cuniculi A genome, increased numbers of affected genes were found among hypothetical genes and genes encoding predicted virulence factors.
Since the divergent proteins comprised mostly those predicted as virulence factors, hypothetical genes with major sequence changes consistently present in all TPE and TPA strains, are candidates for important virulence factors of syphilitic treponemes. This prediction is supported by the relatively high transcription rate of the genes in TPA Nichols during experimental rabbit infections (Šmajs et al., 2005). Interestingly, several of these genes were also found to be altered in T. paraluiscuniculi Cuniculi A, suggesting their role in syphilis pathogenesis (Šmajs et al., 2011).
5. Genetic differences between human T. pallidum subspecies pallidum and the Fribourg-Blanc isolate
Genetic differences between TPA strains and the Fribourg-Blanc isolate have been studied in several previous studies (Grey et al., 2006; Harper et al., 2008a, 2008b) and also in the whole genome fingerprinting study by Mikalová et al. (2010). The Fribourg-Blanc treponeme was isolated in 1962, from a baboon in Guinea, Africa. Although infected baboons showed no signs of infection, the treponemes isolated from these animals were able to infect hamsters (Fribourg-Blanc and Mollaret, 1969). Moreover, experimental infection, using humans, revealed that humans could also be infected (Smith, 1971; Smith et al., 1971). A close relationship between the Fribourg-Blanc treponemes and TPE strains has been found by Gray et al. (2006), based on the phylogeny of the tprC and tprI genes. The WGF study (Mikalová et al., 2010) grouped TPA strains into a separate cluster compared to TPE strains. The Fribourg-Blanc isolate was clustered with TPE strains, although it clustered more distantly than other TPE strains (Mikalová et al., 2010). Interestingly, the Fribourg-Blanc treponemes were shown to contain the largest genome (1140.4 kb) of all uncultivable treponemes characterized on the genome level. However, due to identified duplication of DNA in the intergenic region between TP0696-TP0697 it is unlikely that the Fribourg-Blanc isolate harbors any unique DNA region that is missing in the TPA and TPE strains (Mikalová et al., 2010).
6. Genetic differences among T. pallidum subspecies pallidum strains
Mapping of the genetic diversity among TPA strains is an important step in molecular typing and in epidemiology of syphilitic strains. Genetic differences among TPA type strains were first identified in the tprD gene (Centurion-Lara et al., 2000a) and in the TP0126-TP0127 intergenic region (Marra et al., 2006). In addition to genetic changes among TPA strains, phenotypic differences comprising neuroinvasivity were observed among TPA strains (Tantalo et al., 2005). The WGF analysis revealed several differences among the Nichols, DAL-1, SS14, and Mexico A strains (Mikalová et al., 2010). The estimated sequence identity among TPA strains based on WGF data was 99.92% or higher.
Sequencing of the complete genome of TPA SS14 (Matějková et al., 2008) and the Chicago strain (Giacani et al., 2010) revealed a full list of nucleotide changes among these genomes. When compared to the Nichols strain, sequencing of the SS14 strain revealed 327 single nucleotide changes, 14 deletions and 18 insertions. However, this set excludes changes found in the highly variable tprK gene (Matějková et al., 2008). In general, the observed genetic diversity among TPA strains is similar to the genetic diversity seen in other monomorphic bacterial species, including B. anthracis (Pearson et al., 2004), S. enterica serovar Typhi (Roumagnac et al., 2006), and Y. pestis (Achtman et al., 2004).
Nucleotide diversities computed between pairs of whole genome TPA sequences are shown in Table 2. The diversity between the Nichols and Chicago strains was estimated to more than 150 nt changes and one larger insertion (Giacani et al., 2010). Since this insertion was already identified in a part of Nichols population (Šmajs et al., 2002; Matějková et al., 2008; Mikalová et al., 2010), it does not represent a real difference between both genomes. Moreover, the real number of nucleotide changes differentiating both Nichols and Chicago genomes are likely to be considerably lower (dozens of nucleotide changes) as a result of sequencing errors present in the Nichols genome. The diversity between between SS14 and Mexico A genomes (∼80 nucleotide changes; Pětrošová, unpublished results) is also small, while the diversity between Nichols and SS14 groups (several hundred nucleotide changes) is moderately larger. This clustering was also observed from genome comparisons of 8.3 – 8.4 kb concatenated nucleotide sequences comprising variable chromosomal regions (see Fig. 1B).
Table 2.
Nucleotide diversities between individual pairs of Nichols, SS14, Chicago and Mexico A genomes.
| Genomes | Nucleotide diversity ( ) | Standard deviation of nucleotide diversity |
|---|---|---|
| Nichols*-Chicago | 0.00004 | 0.00002 |
| Nichols*- SS14 | 0.00062 | 0.00031 |
| Nichols*- Mexico A | 0.00072 | 0.00036 |
| SS14-Chicago | 0.00061 | 0.00030 |
| SS14-Mexico A | 0.00015 | 0.00008 |
| Chicago-Mexico A | 0.00068 | 0.00034 |
resequenced version (P. Pospíšilová, unpublished results)
Several studies have mapped the genetic diversity among TPA DNA isolated from clinical material of syphilis patients. The first typing system for detection of genetic diversity (Pillay et al., 1998) revealed 16 subtypes among 46 typeable DNA, isolated from laboratory strains and clinical specimens from the USA, Madagascar, and South Africa, based on the , detected number of repetitions in the arp gene and RFLP polymorphisms in amplified tpr genes. Similar analysis of 45 typeable samples, from Arizona, USA, yielded 10 genotypes (Sutton et al., 2001) and both studies together revealed 22 different TPA subtypes. The work of Pillay et al. (2002) described 35 TPA subtypes among 161 typeable specimens, from South Africa, yielding a total of 44 subtypes identified worldwide. Pope et al. (2005) mapped molecular subtypes among 23 typeable specimens, isolated in North and South Carolina, and identified 7 subtypes. Molepo et al. (2007) identified 4 subtypes in 13 typeable samples from patients with neurosyphilis. Florindo et al. (2008) identified 3 subtypes among 42 typeable specimens from Lisbon, Portugal. Cole et al. (2009) identified 6 subtypes among 58 typeable specimens from Scotland. Martin et al. (2009a) typed 36 samples from Shanghai, China and identified 4 subtypes, and Martin et al. (2010) typed 43 samples in western Canada with four identified subtypes. Altogether, more than 50 individual subtypes in 467 typeable treponemal samples have been isolated from patients. The work of Katz et al. (2010) added to the original CDC typing system (Pillay et al., 1998) analysis of TP0279 (rpsA gene) where the number of repeats in the G homopolymer was used for further typing. Among 69 specimens isolated in San Francisco, 8 individual subtypes were identified.
Moreover, several studies mapped mutations causing macrolide resistance among TPA clinical samples (Katz et al., 2010; Lukehart et al., 2004; Martin et al., 2009a, 2009b; Matějková et al., 2009; Mitchell et al., 2006; Tipple et al., 2011; Van Damme et al., 2009) and an alarming increase in the number of patients infected with TPA strains harboring the A2058G mutation has been detected in USA (Katz et al., 2010; Mitchell et al., 2006). Another mutation (A2059G) leading to macrolide resistance was described recently (Matějková et al., 2009) and a similar frequency of both A2058G and A2059G were found in clinical samples isolated from syphilis patients in the Czech Republic (Woznicová et al., 2010; Flasarová et al., 2011). Both A2058G and A2059G mutations were found also in clinical samples isolated in the USA (Pillay et al., 2011).
Recently, 15 TPA isolates and clinical specimens isolated from 158 syphilis patients in the USA, China, Ireland, and Madagascar were typed with an improved version of the CDC typing system (Marra et al., 2010), which added TP0548 gene sequencing analysis (Flasarová et al., 2006; Woznicová et al., 2007) to the originally described typing system. Compared to the original CDC typing system (Pillay et al., 1998), which identified 14 subtypes, 25 subtypes were identified with the improved version (Marra et al., 2010). As shown with molecular typing studies, genetic diversity within TPA strains is geographically specific, suggesting human population-specific sets of TPA strains.
7. Genetic differences within individual T. pallidum subspecies pallidum strains
To date, several examples of treponemal intrastrain heterogeneity have been published indicating that genetically distinct subpopulations of individual TPA strains exist during infection of human or animal hosts. Stamm and Bergen (2000) identified intrastrain genetic heterogeneity in the tprK of the Nichols and SS14 TPA strains. In addition, variability was also found in the tprJ of the SS14 strain (Stamm and Bergen, 2000). Centurion-Lara et al., (2000a) found intrastrain heterogeneity in the tprK genes of an additional three TPA strains (Sea 81-4, Bal 7, Bal 73-1). tprK sequences found in treponemes isolated from syphilis patients with primary chancres were diverse, but clustered within a sample and not among samples (LaFond et al., 2003). In addition, sequencing of tprK from DNA samples (corresponding to V3-V5 regions of TprK), isolated from patients with syphilis, revealed 48 samples (out of 279) with multiple tprK sequences in one sample (Heymans et al., 2009); indicating that individual patients often contain multiple TPA strains expressing variants of the tprK gene.
Another region with a variable presence of the 1.3-kb DNA sequence was found in the Nichols intergenic region between TP0126 and TP0127. The library of TPA DNA constructed from E. coli contained 6 out of 21 clones with the 1.3 kb insertion and 15 without it (Šmajs et al., 2002). DNA regions corresponding to the 1.3 kb sequence, similar to the tprK gene, were found in all TPA (SS14, DAL-1, Mexico A) and TPE genomes (Samoa D, Gauthier, CDC-2, Fribourg-Blanc) that have been tested (Mikalová et al., 2010).
Except for the variable regions of the tprK gene or tprK-similar sequences, sequencing of the complete TPA SS14 genome has revealed at least 43 nucleotide positions in which the sequence varied within one treponemal strain (Matějková et al., 2008). Multiple intrastrain variants were detected in additional tpr genes (tprC,I,J), in the intergenic region between tprI and tprJ, in TP0402 (flagellum specific ATP synthase), TP0971 (membrane antigen), and TP1029 (hypothetical protein). However, the genome of the SS14 strain was not analyzed systematically for intrastrain heterogeneity and therefore, additional variable genetic loci can be expected (Matějková et al., 2008).
8. Biological relevance of identified genetic changes
Syphilis is a multi-stage disease that is capable of being sexually transmitted, has the potential to invade the central nervous system, and, when untreated in woman, can lead to frequent transplacental congenital infections. In contrast, yaws is a disease with predominant cutaneous or mucocutaneous and bone manifestations (Antal et al., 2002). Although both diseases are distinguished on the basis of epidemiological characteristics and clinical symptoms, these differences do not appear to be fundamental. Yaws can progress to central nervous system and cardiovascular infections as well as to congenital infections (Román and Román, 1986). Moreover, the transmission route of syphilis and yaws appears to reflect opportunity, rather than inherent differences between TPA and TPE strains (Mulligan et al., 2008). Although experimental infection with TPA or TPE strains did not result in complete cross-protection, suggesting differences in the pathogenesis of syphilis and yaws (Miller, 1973; Schell et al., 1982), a similar situation was also found with different TPA strains (Turner and Hollander, 1957). In general, the invasivity of TPE strains appears to be intermediate compared to the more invasive TPA strains and the less invasive T. carateum (Antal et al., 2002).
The small genome size of TPA strains (Fraser et al., 1998; Giacani et al., 2010; Matějková et al., 2008) appears to be the reason for the drastic reduction of treponemal metabolic activities resulting in long generation times (longer than 30 hours; Fieldsteel et al., 1981), sensitivity to oxygen and growth temperature (Stamm et al., 1991) and obligatory host-dependent growth. In contrast to reduced metabolic activities, TPA is an extremely successful pathogen characterized by a low infection dose for humans (as low as about 10 treponemes with a median infection dose of 57 bacteria; Magnuson et al., 1956), the ability to infect any type of human tissue, immune escape resulting in persistence in the host for years or even decades, and the ability to cross the placenta and infect the fetus. Despite efficient host infection, no clearly identified virulence factors have been identified on the basis of sequence analyses of the genomes except for several genes encoding putative hemolysins. However, recombinant expression of these genes in E. coli did not result in a hemolytic phenotype, suggesting another primary role for these genes other than cytolysis (Weinstock et al., 2000b; D. Šmajs, unpublished results). These data indicate the presence of a new, yet unknown, repertoire of genes encoding virulence determinants specific for treponemal/spirochetal pathogens.
8.1. Potential virulence factors determining host specificity of treponemal infections
In general, TPA strains are characterized by low toxicity and high invasiveness, caused, at least to certain extent, by the corkscrew-like motility of treponemes, which allows them to penetrate low viscosity human tissues. Several genes encoding components of treponemal chemotactic sensory systems suggest a possible role for chemotaxis in treponemal pathogenesis.
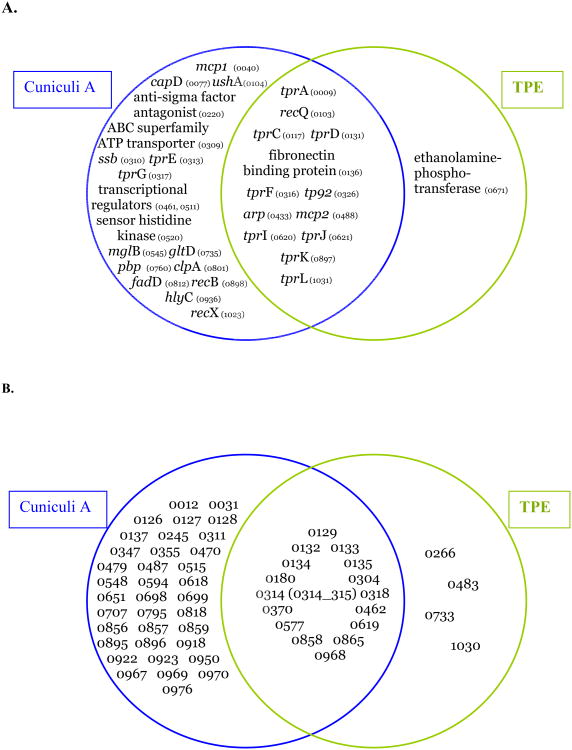
Differences in specific genes between (collectively) the genomes of TPA strains and the Cuniculi A genome are likely to include genes involved in infectivity of TPA strains in humans (for list of genes see Šmajs et al., 2011). In addition, genetic differences between TPA genomes and both the T. paraluiscuniculi and TPE genomes (e.g. tprA,C,D,F,I,J,K,L; recQ; TP0136; tp92, arp; mcp; and hypothetical genes) may be involved in determination of syphilis-specific symptomatology (i.e. invasion of the central nervous system, sexual transmission, and transplacental transmission).
Sequencing of the rabbit pathogen, which is unable to infect humans, has revealed dozens of genes with major sequence changes with the potential to significantly affect their function (Šmajs et al., 2011). Although there is no direct evidence on the role of these genes in treponemal host specificity (Fig. 2), at least three lines of indirect evidence support this prediction: (i) these genes are actively transcribed during experimental rabbit infections (Šmajs et al., 2005, ,2011), (ii) predicted positive evolution of some of these genes in TPE -TPA comparisons (15 genes under positive selection after exclusion of genes with possible recombination events and frameshift mutations (Čejková et al., 2011) and the neutral evolution of several of these genes in TPA – T. paraluiscuniculi comparisons suggesting their importance in human pathogens and their decay in the rabbit pathogen, and (iii) higher numbers of these genes in the group of predicted virulence genes compared to genes encoding components of general metabolism. One of the best studied genes, arp, contains a variable number of repetitive 60 bp sequences in its central region (Pillay et al., 1998) and the variability was also found in the sequence motif (Liu et al., 2007). Moreover, the repetitive motifs of Arp proteins were found to be immunogenic and to contain a fibronectin-binding domain (Liu et al., 2007). Proteins containing tandem repetitions are often outer membrane virulence factors that help the pathogen survive within the host (Denoeud and Vergnaud, 2004). These same proteins have been identified in the genomes of Haemophilus influenzae (Hood et al., 1996), Neisseria meningitidis (Saunders et al., 2000) and Mycoplasma hyorhinis (Citti et al., 1997). Harper et al. (2008b) classified the repetitive motifs of the arp gene into several groups and correlated the variability of repetitive motifs with sexual transmission. Similar variability in the number of invariant repetitions was found in the TP0470 gene (Mikalová et al., 2010; Strouhal et al., 2007). Moreover, the conserved hypothetical protein TP0470 was found to be immunogenic (Brinkman et al., 2006; McKevitt et al., 2005).
Fig. 2.
A. Genes of T. paraluiscuniculi Cuniculi A and TPE with predicted cell function with major sequence changes compared to the Nichols orthologs. Cuniculi A genes containing internal frameshifts and/or major sequence changes (Šmajs et al., 2011) and TPE genes encoding proteins with six or more amino acid changes and/or major sequence changes when compared to TPA genes (Čejková et al., 2011). B. Genes of T. paraluiscuniculi Cuniculi A and TPE with unknown function with major sequence changes compared to the Nichols orthologs. Same criteria were used as in panel A.
8.2 Potential virulence factors determining invasivity of treponemal infections
Genes specifically affected in TPE genomes (TP0671 encoding ethanolamine phosphotransferase, hypothetical genes) could code for factors increasing invasivity of TPA strains (Fig. 2). Except for the tpr genes, only TP0077, TP0326, TP0520, and TP0936 were among the predicted virulence factors on a list containing 67 possible virulence factors (Weinstock et al., 1998). This discrepancy likely reflects our insufficient understanding of the pathogenesis of TPA and TPE infections. The C. jejuni phosphoethanolamine transferase modifies the lipooligosaccharide lipid anchor (lipid A) and the flagellar rod protein, FlgG (Cullen and Trent, 2010). Since treponemes do not contain lipopolysaccharides, it may be involved in the modification of the proteins encoded by the flgG-1 and flgG-2 genes (TP0960, TP0961) that are present in both syphilis and yaws treponemes.
8.3 Positive selection of treponemal genes
The genomic data provides sequence information that can be used for prediction of genes involved in pathogenesis by estimation of selection type. The type of selection has been predicted for several treponemal genes based on available sequences from T. paraluiscuniculi, TPE and TPA strains. As shown in Table 3, a positive selection type was identified between several orthologous sequences. The predicted type of selection together with an assumption that T. paraluiscuniculi is a descendant of TPA or TPE strains in the process of adaptation to rabbits (Šmajs et al., 2011), can be used for identification of genes involved in the pathogenesis of particular treponemal diseases. The tp92 (TP0326) gene is under positive selective pressure (Table 3) in TPE and TPA treponemes and therefore has a potential role in syphilis and/or yaws pathogenesis. In contrast, this gene in the Cuniculi A strain is under purifying (negative) selection, indicating its important cellular function but no major role in adaptation to rabbits or immune escape during rabbit syphilis. In contrast, the arp (TP0433) gene appears to have been positively selected in the Cuniculi A strain (Harper et al., 2008b) compared to TPA/TPE strains, indicating its possible role in rabbit syphilis. The mcp (TP0488) gene appears to be positively selected in TPA – TPE comparison, suggesting a role in adaptation of the yaws and/or syphilis treponemes.
Table 3.
Three selected genes with predicted type of selection derived from orthologous gene sequence comparisons.
| Gene | Type of predicted selection* between (no. of polymorphic sites; statistical sign ificance) | |||||
|---|---|---|---|---|---|---|
| Cuniculi A/ Nichols | Cuniculi A/ Samoa D | Cuniculi A/ SS14 | Samoa D/ Nichols | Samoa D/ SS14 | SS14/ Nichols | |
| tp92 | purifying | purifying | purifying | positive | positive | |
| (TP0326) | (57; p=0.017) | (56; p=0.008) | (63; p=0.037) | (10; p=0.502) | (21; p=0.047) | (12; p=0.037) |
| arp | positive | positive | positive | positive | positive | ** |
| (TP0433) | (73; p=0.000) | (58; p=0.000) | (75; p=0.000) | (7; p=0.015) | (11; p=0.008) | (10; p=0.325) |
| mcp | positive | positive | positive? | |||
| (TP0488) | (71; p=0.153) | (55; p=0.557) | (68; p=0.142) | (34; p=0.000) | (31; p=0.015) | (4; NA) |
neutral selection not shown;
TPA strains (and clinical isolates) are known to differ in the number of repetitions in the arp genes (Pillay et al., 1998; Pope et al., 2005; Molepo et al., 2007; Florindo et al., 2008; Cole et al., 2009; Martin et al., 2009a; Marra et al., 2010). Positive selection thus appears to operate on a repetition number level.
NA, not applicable
Of 78 investigated TPE divergent genes, positive selection in 15 orthologous genes has been predicted. The positively selected genes were often found to encode exported or membrane proteins including predicted lipoproteins and outer membrane proteins suggesting that these proteins may be responsible for differences in pathogenesis between syphilis and yaws (Čejková et al., 2011). Predicted positive selection in metabolic, transport, and cell- process genes suggest that positive selection of these genes could be correlated to adaptive processes, such as climate adaptation of treponemes.
8.4 Lack of intrastrain adaptive mutations
Sequencing of the Cuniculi A genome revealed gene fusions compared to the originally annotated Nichols genome (Fraser et al., 1998) where 52 Nichols orthologs were fused into 25 genes in the Cuniculi A genome (Šmajs et al., 2011). Ongoing resequencing of the Nichols genome (P. Pospíšilová, personal communication) has revealed that most of the observed gene fusions are also present in the Nichols genome. Since the Nichols strain was originally isolated in 1912 from a patient with neurosyphilis (Nichols and Hough, 1913) and, since then, propagated in rabbits, questions regarding newly emerged, adaptive, nucleotide changes important for rabbit infectivity have appeared. Despite extended propagation in rabbits, the Nichols strain is still virulent as demonstrated by human inoculations (Magnuson et al., 1956) as well as accidental infections of laboratory personnel (Chacko, 1966; Fitzgerald et al., 1976). Since whole genome sequencing of the Nichols strain (Fraser et al., 1998) and Nichols resequencing (Nichols 4787, S.J. Norris) used different Nichols preparations, we examined highly related TPA genomes (Chicago, DAL-1, and Mexico A) for the presence of the newly identified changes (Šmajs et al., 2011). Since similar sets of nucleotide changes were also found in TPA Chicago (Giacani et al., 2010) and the preliminary DAL-1 and Mexico A genomes (unpublished data), the potential presence of recently emerged intrastrain adaptive mutations in the Nichols genome was excluded. Instead, the observed changes represent sequencing errors in the published Nichols genome (Fraser et al., 1998; Šmajs et al., 2011).
8.5 Variability in tpr genes and their possible function in treponemal infections
In treponemal genomes, there are 12 paralogous genes (or pseudogenes) classified into 3 subfamilies (subfamily I, tprC,D,F,I; II, tprE,G,J; and III, tprA,B,H,K,L) Several of these proteins, belonging to all three subfamilies, have been recently predicted as outer membrane proteins (TprB,C,D,E,F,I,J; Cox et al., 2010). In addition to these proteins, Centurion-Lara et al. (1999) showed that tprK encodes an outer membrane protein, although others have questioned this finding (Hazlett et al., 2001). Recently, Giacani et al. (2010) showed that the host immune response selects new TprK variants during experimental infection which is consistent with the surface localization of TprK. Subfamily II tpr genes are expressed differently in different TPA isolates with G homopolymers of variable length in the promoter regions (Giacani et al., 2007, 2009). Since most of Tpr proteins elicit an antibody response and expression of tpr genes are time and strain specific, their role in persistence of treponemal infections in immunocompetent host has been suggested (Leader et al., 2003). TprK protein is the most variable treponemal protein having several variants within one strain of TPA (except for the rabbit propagated Nichols strain; LaFond et al., 2006). Moreover, it has been shown that most variable regions of TprK showed increased sequence diversity upon rabbit experimental infection (La Fond et al., 2003). The extreme variability of the tprK gene is generated through a gene conversion mechanism (Centurion-Lara et al., 2004) causing TprK to undergo antigenic variation, which may in turn promote chronic infection. Whereas T-cells recognize conserved regions of TprK, antibodies are directed against the variable regions (Morgan et al., 2002a). Immunization with recombinant Nichols TprK led to partial protection against a Nichols challenge, but provided less protection against TPA with heterologous TprK (Centurion-Lara et al., 1999; Morgan et al., 2002b, 2003). The fact that tpr genes are the most variable gene family in treponemal strains, suggests (Centurion-Lara et al., 2000a, 2000b) their role in pathogenesis of treponematoses as well as in host specificity. The variability of tprK appears to be one of principal mechanisms of immune evasion allowing reinfection of hosts that had already been previously infected (Palmer et al., 2009).
9. Origin of syphilis and evolution of pathogenic treponemes
Most of the work on syphilis origin and evolution is based on paleontological findings; however, several studies have already started to use genetic data. The work of Gray et al., (2006) analyzed 6 tpr genes in TPA, TPE, TEN, the Fribourg-Blanc isolate and T. paraluiscuniculi strains and identified intragenomic recombination as an important mechanism of tpr gene evolution. Moreover, this work does not support the origin of TPA strains in recent history (less than 500 years). The work performed by Harper et al. (2008a) involved genetic analyses of 21 chromosomal regions of several (26) treponemal strains and isolates. This work proposes that T. pallidum as a species originated in the Old World as a non-venereal infection and then spread to the New World as yaws. Consequently, treponemal strains from the Americas were introduced to Europe and became the syphilis-causing strain. However, evolutionary order with respect to TPA and TPE strains cannot be unambiguously inferred from this analysis (Mulligan et al., 2008), especially in the light of genome decay identified in T. paraluiscuniculi (Šmajs et al., 2011), which suggests that T. paraluiscuniculi is a descendant of an ancestor of pallidum and pertenue strains rather than the opposite. Measurement of evolution rate in treponemes and its comparison with paleopathological evidence and with evolution rates of other bacteria is consistent with the divergence of syphilis from other human treponematoses 5 - 16.5 thousand years ago (de Melo et al., 2010).
Historically, there have been several hypotheses on the origin of syphilis, including the Columbian, Pre-Columbian, Alternative and the Unitarian hypotheses. Whereas the Columbian hypothesis presumed the existence of syphilis in America with subsequent import by Columbus' crew into Europe (Mays et al., 2003; Naranjo, 1994), the Pre-Columbian hypothesis assumed the existence of syphilis and other treponematoses in the Old and New World a long time before Columbus' era and the misidentification of pre-Columbian syphilis cases as leprosy or other diseases (Meyer et al., 2002; Powel and Cook, 2005). According to the Unitarian hypothesis, all treponemal diseases (syphilis, yaws, bejel, and pinta) are caused by the identical microorganism and disease symptomatology reflects differences in climate and social factors (Powel and Cook, 2005). The Alternative theory suggests trans-species transmission of syphilitic treponemes from African baboons (Livingstone, 1991).
Based on consistent genetic differences found between strains causing yaws and syphilis (Gray et al., 2006; Harper et al., 2008a, 2008b; Čejková et al., 2011), the Unitarian hypothesis can be rejected. The genetic distance between TPA and TPE strains appears to be inconsistent with a recent emergence of syphilis in the 15th century and TPA strains appear to be at least several thousand years old (de Melo et al., 2010; Gray et al., 2006). However, one cannot exclude a very rapid adaptation of a parasite to its host under the right circumstances.
Since bone alterations caused by syphilis, yaws and bejel are distinctive for these diseases (Rothschild, 2005) and the osseotype of syphilis in pre-Columbian era was found in the New World but not in Europe, Africa or Asia (Rothschild, 2005; Rothschild and Rothschild, 2000), transmission of syphilis by Columbus' crew into Europe is a plausible explanation. However, other authors suggest the presence of syphilis in other parts of the world, in addition to the New World, in pre-Columbian era (de Melo et al., 2010). Support for the Alternative hypothesis (Livingstone, 1991) comes from the fact that concurrently with Columbus' expeditions, exploration of the African continent was also intensifying, which could have increased exposure to a possible reservoir of yaws treponemes in the human (and primate) populations of Africa.
Although our knowledge regarding the evolution of syphilis treponemes is rather fragmentary, there are several points revealed by genetic and genomic studies: (i) there is a close genetic relationship between the Fribourg-Blanc isolate and yaws strains (Gray et al., 2006; Harper et al., 2008a; Mikalová et al., 2010) as well as between treponemes causing infections in wild baboons and TPE strains (Knauf et al., 2011), which indicates a common origin of these treponemes. Moreover, the Fribourg-Blanc treponeme has the largest genome amongst the investigated TPA and TPE strains (Mikalová et al., 2010), (ii) genetic diversity between TPA and TPE strains is consistent with TPA evolution of several thousand years (de Melo et al., 2010; Gray et al., 2006), and (iii) pathogenic treponemes evolve by decreasing genome size while adapting to their hosts (Šmajs et al., 2011). The available genetic and genomic data are thus inconsistent with any of the remaining theories regarding the origin of syphilis, but may combine some aspects of all of them in the origin of the African baboon treponemal strains, in evolution of TPA strains for several thousand years and in sudden import of TPA strains from the New World by Columbus' crew into Europe.
10. Targets for molecular detection and typing of TPA and TPE strains
Except for tpr genes and the arp gene, TP0136, TP0548, TP0326 and TP0488 genes were identified as those with the greatest nucleotide differences among TPA strains tested, including Nichols, SS14, DAL-1, Mexico A, Grady, MN-3, Philadelphia-1, Philadelphia-2, and Bal-73-01 (D. Šmajs, unpublished results). All the investigated strains clustered in two subclusters containing either the Nichols or the SS14 strain (Fig. 1B). Together with the 23S rDNA locus, the TP0136 and TP0548 have already been tested for typing of syphilis causing strains isolated in the Czech Republic. Out of typeable treponemal DNA samples taken from 64 patients, nine different genotypes were identified (Flasarová et al., 2011). More importantly, the identified genotypes were found to independently combine with each other and also with the number of repetitions in the arp gene and the restriction profile of tprEGJ genes (Pillay et al., 1998). The recently improved CDC typing system combines arp and tprEGJ gene typing (Pillay et al., 1998) with analysis of rpsA gene G repeats (TP0279) (Katz et al., 2010) or with sequencing of part of the TP0548 gene (Marra et al., 2010). Since human populations are partially separated based on geographic origin, religion, native language, etc. (e.g. T. pallidum DNA from patients in Madagascar shared only one out of six subtypes identical to those isolated from the USA patients, Marra et al., 2010), it is likely that population-specific typing systems would be needed for precise epidemiological mapping of syphilis.
Whole genome analyses of TPE strains (Mikalová et al., 2010) and subsequent whole genome sequencing (Čejková et al., 2011) revealed several regions suitable for a molecular diagnosis of the yaws causing strains including TP0266 and TP0316 genes, intergenic region (IGR) between TP0548-TP0549, and TP1030-TP1031 genes. The observed indels ranged between 33-635 nt in length and can be found in the Samoa D genome (GenBank acc. no.: CP002374). However, these regions need to be analyzed in a larger number of TPE strains. Interestingly, all of these indels were also found in the Fribourg-Blanc genome. However, several specific indels differentiate the Fribourg-Blanc isolate from other individual TPE strains (Mikalová et al., 2010). Whole genome sequencing of the TEN strain, Bosnia A, that is in progress (D. Šmajs, unpublished data) has already revealed indels in IGR TP0085-TP0086, and in genes TP0136, TP0326, and TP0865, ranging between 13 and 0.06 kbp, which differentiate the bejel treponeme from both the TPA and TPE strains. Further work will test TPE and TEN specific molecular signatures as targets for specific molecular detection of yaws and bejel treponemes.
11. Conclusions
We are still some distance from understanding of the genetic basis of syphilis pathogenesis and of the strategies that T. pallidum uses to survive, invade and adapt in human hosts, especially when we consider its ability to cause, when untreated, life-long infections and the ability to infect all human tissue types. In spite of our relative lack of understanding with regard to the pathogenesis of T. pallidum, it is nonetheless easy to treat T. pallidum infections with antibiotics. Because of this and other features, such as a relatively short incubation period, it should be theoretically possible to eradicate syphilis (Rompalo et al., 2001) in the relatively near future. Future will reveal if the discovery of T. pallidum molecular cell strategies in pathogenesis of human infection will come sooner than the syphilis eradication or vice versa.
>We reviewed genetic diversity in pathogenic uncultivable treponemes.
>Genes involved in human infectivity and treponemal invasivity were discussed.
>Variability within syphilis strains was considered in the light of molecular epidemiology.
>Treponemal evolution was confronted with genetic data.
Acknowledgments
This work was supported by the grant of the Grant Agency of the Czech Republic (310/04/0021) and the grant of the Ministry of Health of the Czech Republic (NT11159-5/2010) and Ministry of Education of the Czech Republic (VZ MSM0021622415) to D.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal GM, Lukehart SA, Meheus AZ. The endemic treponematoses. Microbes Infect. 2002;4:83–94. doi: 10.1016/s1286-4579(01)01513-1. [DOI] [PubMed] [Google Scholar]
- Brinkman MB, McGill MA, Pettersson J, Rogers A, Matějková P, Šmajs D, Weinstock GM, Norris SJ, Palzkill T. A novel Treponema pallidum antigen, TP0136, is an outer membrane protein that binds human fibronectin. Infect Immun. 2008;76:1848–1857. doi: 10.1128/IAI.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman MB, McKevitt M, McLoughlin M, Perez C, Howell J, Weinstock GM, Norris SJ, Palzkill T. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J Clin Microbiol. 2006;44:888–891. doi: 10.1128/JCM.44.3.888-891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Castro C, Lukehart SA, Van Voorhis WC. Sequence conservation of glycerophosphodiester phosphodiesterase among Treponema pallidum strains. Infect Immun. 1999;67:3168–3170. doi: 10.1128/iai.67.6.3168-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron CE, Lukehart SA, Castro C, Molini B, Godornes C, Van Voorhis WC. Opsonic potential, protective capacity, and sequence conservation of the Treponema pallidum subspecies pallidum Tp92. J Infect Dis. 2000;181:1401–1413. doi: 10.1086/315399. [DOI] [PubMed] [Google Scholar]
- Castellani A. Further observations on parangi (Yaws) Brit Med J. 1905;1905:1330–1331. doi: 10.1136/bmj.2.2342.1330-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čejková D, Zobaníková M, Chen L, Pospíšilová P, Strouhal M, Qin X, Mikalová L, Norris SJ, Muzny DM, Gibbs RA, Fulton LL, Sodergren E, Weinstock GM, Šmajs D. Whole genome sequences of three Treponema pallidum ssp. pertenue strains: yaws and syphilis treponemes differ in less than 0.2% of the genome sequence. PLo S Neglect Trop D. 2011 doi: 10.1371/journal.pntd.0001471. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, Castro C, Barrett L, Cameron C, Mostowfi M, Van Voorhis WC, Lukehart SA. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J Exp Med. 1999;189:647–656. doi: 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, Castro C, Castillo R, Shaffer JM, Van Voorhis WC, Lukehart SA. The flanking region sequences of the 15-kDa lipoprotein gene differentiate pathogenic treponemes. J Infect Dis. 1998;177:1036–1040. doi: 10.1086/515247. [DOI] [PubMed] [Google Scholar]
- Centurion-Lara A, Godornes C, Castro C, Van Voorhis WC, Lukehart SA. The tprK gene is heterogenous among Treponema pallidum strains and has multiple alleles. Infect Immun. 2000b;68:824–831. doi: 10.1128/iai.68.2.824-831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, LaFond RE, Hevner K, Godornes C, Molini BJ, Van Voorhis WC, Lukehart SA. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol Microbiol. 2004;52:1579–1596. doi: 10.1111/j.1365-2958.2004.04086.x. [DOI] [PubMed] [Google Scholar]
- Centurion-Lara A, Molini BJ, Godornes C, Sun E, Hevner K, Van Voorhis WC, Lukehart SA. Molecular differentiation of Treponema pallidum subspecies. J Clin Microbiol. 2006;44:3377–3380. doi: 10.1128/JCM.00784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, Sun ES, Barrett LK, Castro C, Lukehart SA, Van Voorhis WC. Multiple alleles of Treponema pallidum repeat gene D in Treponema pallidum isolates. J Bacteriol. 2000a;182:2332–2335. doi: 10.1128/jb.182.8.2332-2335.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko CW. Accidental human infection in the laboratory with Nichols rabbit-adapted virulent strain of Treponema pallidum. Bull World Health Organ. 1966;35:809–810. [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chi KH, Pillay A, Nachamkin E, Ballard R. Detection of the 23S rRNA point mutations (A2058G and A2059G) associated with azithromycin resistance in Treponema pallidum using a TaqMan-based real-time Triplex-PCR assay. Sex Transm Dis. 2011;87(Suppl):A298. doi: 10.1128/JCM.02770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citti C, Kim MF, Wise KS. Elongated versions of Vlp surface lipoproteins protect Mycoplasma hyorhinis escape variants from growth-inhibiting host antibodies. Infect Immun. 1997;65:1773–1785. doi: 10.1128/iai.65.5.1773-1785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MJ, Chisholm SA, Palmer HM, Wallace LA, Ison CA. Molecular epidemiology of syphilis in Scotland. Sex Transm Infect. 2009;85:447–451. doi: 10.1136/sti.2009.036301. [DOI] [PubMed] [Google Scholar]
- Cox DL, Luthra A, Dunham-Ems S, Desrosiers DC, Salazar JC, Caimano MJ, Radolf JD. Surface immunolabeling and consensus computational framework to identify candidate rare outer membrane proteins of Treponema pallidum. Infect Immun. 2010;78:5178–5194. doi: 10.1128/IAI.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci USA. 2010;107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo FL, de Mello JC, Fraga AM, Nunes K, Eggers S. Syphilis at the crossroad of phylogenetics and paleopathology. PLoS Negl Trop Dis. 2010;4:e575. doi: 10.1371/journal.pntd.0000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud F, Vergnaud G. Identification of polymorphic tandem repeats by direct comparison of genome sequence from different bacterial strains: a web-based resource. BMC Bioinformatics. 2004;5:4. doi: 10.1186/1471-2105-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiacomo RF, Lukehart SA, Talburt CD, Baker-Zander SA, Condon J, Brown CW. Clinical course and treatment of venereal spirochaetosis in New Zealand white rabbits. Br J Vener Dis. 1984;60:214–218. doi: 10.1136/sti.60.4.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiacomo RF, Lukehart SA, Talburt CD, Baker-Zander SA, Giddens WE, Jr, Condon J, Brown CW. Chronicity of infection with Treponema paraluis-cuniculi in New Zealand white rabbits. Genitourin Med. 1985;61:156–164. doi: 10.1136/sti.61.3.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiacomo RF, Talburt CD, Lukehart SA, Baker-Zander SA, Condon J. Treponema paraluis-cuniculi infection in a commercial rabbitry: epidemiology and serodiagnosis. Lab Anim Sci. 1983;33:562–566. [PubMed] [Google Scholar]
- Dos Vultos T, Mestre O, Rauzier J, Golec M, Rastogi N, Rasolofo V, Tonjum T, Sola C, Matic I, Gicquel B. Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS One. 2008;2:e1538. doi: 10.1371/journal.pone.0001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieldsteel AH, Cox DL, Moeckli RA. Cultivation of virulent Treponema pallidum in tissue culture. Infect Immun. 1981;32:908–915. doi: 10.1128/iai.32.2.908-915.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald JJ, Johnson RC, Smith M. Accidental laboratory infection with Treponema pallidum, Nichols strain. J Am Vener Dis Assoc. 1976;3:76–78. [PubMed] [Google Scholar]
- Flasarová M, Pospíšilová P, Mikalová L, Vališová Z, Dastychová E, Strnadel R, Kuklová I, Woznicová V, Zákoucká H, Šmajs D. Sequencing-based molecular typing of Treponema pallidum strains in the Czech Republic: all identified genotypes are related to the sequence of the SS14 strain. Acta Derm Venereol. 2011 doi: 10.2340/00015555-1335. accepted. [DOI] [PubMed] [Google Scholar]
- Flasarová M, Šmajs D, Matějková P, Woznicová V, Heroldová-Dvořáková M, Votava M. Molecular detection and subtyping of Treponema pallidum subsp pallidum in clinical specimens. Epidemiol Mikrobiol Imunol. 2006;55:105–111. [PubMed] [Google Scholar]
- Florindo C, Reigado V, Gomes JP, Azevedo J, Santo I, Borrego MJ. Molecular typing of Treponema pallidum clinical strains from Lisbon, Portugal. J Clin Microbiol. 2008;46:3802–3803. doi: 10.1128/JCM.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, MacDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith HO, Venter JC. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Fribourg-Blanc A, Mollaret HH. Natural treponematotis of the African primate. Primates Med. 1969;3:113–121. [PubMed] [Google Scholar]
- Gastinel P, Vaisman A, Hamelin A, Dunoyer F. Study of a recently isolated strain of Treponema pertenue. Ann Dermatol Syphiligr Paris. 1963;90:155–161. [PubMed] [Google Scholar]
- Giacani L, Godornes C, Puray-Chavez M, Guerra-Giraldez C, Tompa M, Lukehart SA, Centurion-Lara A. TP0262 is a modulator of promoter activity of tpr Subfamily II genes of Treponema pallidum ssp. pallidum. Mol Microbiol. 2009;72:1087–1099. doi: 10.1111/j.1365-2958.2009.06712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacani L, Jeffrey BM, Molini BJ, Le HT, Lukehart SA, Centurion-Lara A, Rockey DD. Complete genome sequence and annotation of the Treponema pallidum subsp pallidum Chicago strain. J Bacteriol. 2010;192:2645–2646. doi: 10.1128/JB.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacani L, Lukehart S, Centurion-Lara A. Length of guanosine homopolymeric repeats modulates promoter activity of subfamily II tpr genes of Treponema pallidum ssp. pallidum. FEMS Immunol Med Microbiol. 2007;51:289–301. doi: 10.1111/j.1574-695X.2007.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacani L, Molini BJ, Kim EY, Godornes BC, Leader BT, Tantalo LC, Centurion-Lara A, Lukehart SA. Antigenic variation in Treponema pallidum: TprK sequence diversity accumulates in response to immune pressure during experimental syphilis. J Immunol. 2010;184:3822–3829. doi: 10.4049/jimmunol.0902788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacani L, Sun ES, Hevner K, Molini BJ, Van Voorhis WC, Lukehart SA, Centurion-Lara A. Tpr homologs in Treponema paraluiscuniculi Cuniculi A strain. Infect Immun. 2004;72:6561–6576. doi: 10.1128/IAI.72.11.6561-6576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Spratt BG. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves S, Downes J. Experimental infection of man with rabbit-virulent Treponema paraluis-cuniculi. Br J Vener Dis. 1981;57:7–10. doi: 10.1136/sti.57.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RR, Mulligan CJ, Molini BJ, Sun ES, Giacani L, Godornes C, Kitchen A, Lukehart SA, Centurion-Lara A. Molecular evolution of the tprC, D, I, K, G, and J genes in the pathogenic genus Treponema. Mol Biol Evol. 2006;23:2220–2233. doi: 10.1093/molbev/msl092. [DOI] [PubMed] [Google Scholar]
- Harper KN, Liu H, Ocampo PS, Steiner BM, Martin A, Levert K, Wang D, Sutton M, Armelagos GJ. The sequence of the acidic repeat protein (arp) gene diferentiates venereal from nonvenereal Treponema pallidum subspecies, and the gene has evolved under strong positive selection in the subspecies that causes syphilis. FEMS Immunol Med Microbiol. 2008b;53:322–332. doi: 10.1111/j.1574-695X.2008.00427.x. [DOI] [PubMed] [Google Scholar]
- Harper KN, Ocampo PS, Steiner BM, George RW, Silverman MS, Bolotin S, Pillay A, Saunders NJ, Armelagos GJ. On the origin of the treponematoses: a phylogenetic approach. PloS Negl Trop Dis. 2008a;2:e148. doi: 10.1371/journal.pntd.0000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett KR, Sellati TJ, Nguyen TT, Cox DL, Clawson ML, Caimano MJ, Radolf JD. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J Exp Med. 2001;193:1015–1026. doi: 10.1084/jem.193.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymans R, Kolader ME, van der Helm JJ, Coutinho RA, Bruisten SM. TprK gene regions are not suitable for epidemiological syphilis typing. Eur J Clin Microbiol Infect Dis. 2009;28:875–878. doi: 10.1007/s10096-009-0717-5. [DOI] [PubMed] [Google Scholar]
- Hood DW, Deadman ME, Jennings MP, Bisercic M, Fleischmann RD, Venter JC, Moxon ER. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci U S A. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsthal E. Untersuchungen über eine syphilisähnliche Spontanerkrankungen des Kaninchens (Paralues cuniculi) Derm Wschr. 1920;71:569–571. [Google Scholar]
- Katz KA, Pillay A, Ahrens K, Kohn RP, Hermanstyne K, Bernstein KT, Ballard RC, Klausner JD. Molecular epidemiology of syphilis - San Francisco, 2004-2007. Sex Transm Dis. 2010;37:660–663. doi: 10.1097/OLQ.0b013e3181e1a77a. [DOI] [PubMed] [Google Scholar]
- Kidgell C, Reichard U, Wain J, Linz B, Torpdahl M, Dougan G, Achtman M. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect Genet Evol. 2002;2:39–45. doi: 10.1016/s1567-1348(02)00089-8. [DOI] [PubMed] [Google Scholar]
- Knauf S, Batamuzi EK, Mlengeya T, Kilewo M, Lejora IA, Nordhoff M, Ehlers B, Harper KN, Fyumagwa R, Hoare R, Failing K, Wehrend A, Kaup FJ, Leendertz FH, Mätz-Rensing K. Treponema Infection Associated With Genital Ulceration in Wild Baboons. Vet Pathol. 2011 Mar 16;2011 doi: 10.1177/0300985811402839. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB. Bergey's Manual of Systematic Bacteriology. second. Springer; New York: 2010. pp. 501–531. [Google Scholar]
- LaFond RE, Centurion-Lara A, Godornes C, Rompalo AM, Van Voorhis WC, Lukehart SA. Sequence diversity of Treponema pallidum subsp. pallidum tprK in human syphilis lesions and rabbit-propagated isolates. J Bacteriol. 2003;185:6262–6268. doi: 10.1128/JB.185.21.6262-6268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFond RE, Centurion-Lara A, Godornes C, Van Voorhis WC, Lukehart SA. TprK sequence diversity accumulates during infection of rabbits with Treponema pallidum subsp. pallidum Nichols strain Infect Immun. 2006;74:1896–1906. doi: 10.1128/IAI.74.3.1896-1906.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader BT, Hevner K, Molini BJ, Barrett LK, Van Voorhis WC, Lukehart SA. Antibody responses elicited against the Treponema pallidum repeat proteins differ during infection with different isolates of Treponema pallidum subsp. pallidum. Infect Immun. 2003;71:6054–6057. doi: 10.1128/IAI.71.10.6054-6057.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levaditi C, Marie A, Nicolau S. Virulence pour l'homme du spirochète de la spirillose spontanée du lapin. C R Acad Sci. 1921;172:1542–1543. [Google Scholar]
- Liska SL, Perine PL, Hunter EF, Crawford JA, Feelez JC. Isolation and transportation of Treponema pertenue in golden hamsters. Curr Microbiol. 1982;7:41–43. [Google Scholar]
- Liu H, Rodes B, George R, Steiner B. Molecular characterization and analysis of a gene encoding the acidic repeat protein (Arp) of Treponema pallidum. J Med Microbiol. 2007;56:715–721. doi: 10.1099/jmm.0.46943-0. [DOI] [PubMed] [Google Scholar]
- Livingstone FB. On the origin of syphilis: an alternative hypothesis. Curr Anthropol. 1991;32:587–590. [Google Scholar]
- Lukehart SA, Godornes C, Molini BJ, Sonnett P, Hopkins S, Mulcahy F, Engelman J, Mitchell SJ, Rompalo AM, Marra CM, Klausner JD. Macrolide resistance in Treponema pallidum in the United States and Ireland. N Engl J Med. 2004;351:154–158. doi: 10.1056/NEJMoa040216. [DOI] [PubMed] [Google Scholar]
- MacDougall J, Saint Girons I. Physical map of the Treponema denticola circular chromosome. J Bacteriol. 1995;177:1805–1811. doi: 10.1128/jb.177.7.1805-1811.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson HJ, Thomas EW, Olansky S, Kaplan BI, de Mello L, Cutler JC. Inoculation syphilis in human volunteers. Medicine (Baltimore) 1956;35:33–82. doi: 10.1097/00005792-195602000-00002. [DOI] [PubMed] [Google Scholar]
- Marra CM, Colina AP, Godornes C, Tantalo LC, Puray M, Centurion-Lara A, Lukehart SA. Antibiotic selection may contribute to increases in macrolide-resistant Treponema pallidum. J Infect Dis. 2006;194:1771–1773. doi: 10.1086/509512. [DOI] [PubMed] [Google Scholar]
- Marra CM, Sahi SK, Tantalo LC, Godornes C, Reid T, Behets F, Rompalo A, Klausner JD, Yin YP, Mulcahy F, Golden MR, Centurion-Lara A, Lukehart SA. Enhanced molecular typing of Treponema pallidum: geographical distribution of strain types and association with neurosyphilis. J Infect Dis. 2010;202:1380–1388. doi: 10.1086/656533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin IE, Gu W, Yang Y, Tsang RSW. Macrolide resistance and molecular types of Treponema pallidum causing primary syphilis in Shanghai, China. Clin Infect Dis. 2009a;49:515–521. doi: 10.1086/600878. [DOI] [PubMed] [Google Scholar]
- Martin IE, Tsang RS, Sutherland K, Anderson B, Read R, Roy C, Yanow S, Fonseca K, White W, Kandola K, Kouadjo E, Singh AE. Molecular typing of Treponema pallidum strains in western Canada: predominance of 14d subtypes. Sex Transm Dis. 2010;37:544–548. doi: 10.1097/OLQ.0b013e3181d73ce1. [DOI] [PubMed] [Google Scholar]
- Martin IE, Tsang RSW, Sutherland K, Tilley P, Read R, Anderson B, Roy C, Singh AE. Molecular characterization of syphilis in pacients in Canada: azitromycin resistance and detection of Treponema pallidum DNA in whole-blood samples versus ulcerative swabs. J Clin Microbiol. 2009b;47:1668–1673. doi: 10.1128/JCM.02392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matějková P, Flasarová M, Zákoucká H, Bofek M, Kfemenová S, Arenberger P, Woznicová V, Weinstock GM, Šmajs D. Macrolide treatment failure in a case of secondary syphilis: a novel A2059G mutation in the 23S rRNA gene of Treponema pallidum subsp. pallidum. J Med Microbiol. 2009;58:832–836. doi: 10.1099/jmm.0.007542-0. [DOI] [PubMed] [Google Scholar]
- Matějková P, Strouhal M, Šmajs D, Norris SJ, Palzkill T, Petrosino JF, Sodergren E, Norton JE, Singh J, Richmond TA, Molla MN, Albert TJ, Weinstock GM. Complete genome sequence of Treponema pallidum ssp. pallidum strain SS14 determined with oligonucleotide arrays. BMC Microbiol. 2008;8:76. doi: 10.1186/1471-2180-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays S, Crane-Kramer G, Bayliss A. Two probable cases of treponemal disease of Medieval date from England. Am J Phys Anthropol. 2003;120:133–143. doi: 10.1002/ajpa.10132. [DOI] [PubMed] [Google Scholar]
- McGill MA, Edmondson DG, Carroll JA, Cook RG, Orkiszewski RS, Norris SJ. Characterization and serologic analysis of the Treponema pallidum proteome. Infect Immun. 2010;78:2631–2643. doi: 10.1128/IAI.00173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKevitt M, Brinkman MB, McLoughlin M, Perez C, Howell JK, Weinstock GM, Norris SJ, Palzkill T. Genome scale identification of Treponema pallidum antigens. Infect Immun. 2005;73:4445–4450. doi: 10.1128/IAI.73.7.4445-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKevitt M, Patel K, Šmajs D, Marsh M, McLoughlin M, Norris SJ, Weinstock GM, Palzkill T. Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res. 2003;13:1665–1674. doi: 10.1101/gr.288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Jung C, Kohl T, Poenicke A, Poppe A, Alt KW. Syphilis 2001--a palaeopathological reappraisal. Homo. 2002;53:39–58. doi: 10.1078/0018-442x-00037. [DOI] [PubMed] [Google Scholar]
- Miao RM, Fieldsteel AH. Genetics of Treponema: relationship between Treponema pallidum and five cultivable treponemes. J Bacteriol. 1978;133:101–107. doi: 10.1128/jb.133.1.101-107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao RM, Fieldsteel AH. Genetic relationship between Treponema pallidum and Treponema pertenue, two noncultivable human pathogens. J Bacteriol. 1980;141:427–429. doi: 10.1128/jb.141.1.427-429.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikalová L, Strouhal M, Čejková D, Zobaníková M, Pospíšilová P, Norris SJ, Sodergren E, Weinstock GM, Šmajs D. Genome analysis of Treponema pallidum subsp. pallidum and subsp pertenue strains: most of the genetic differences are localized in six regions. PLoS One. 2010;5:e15713. doi: 10.1371/journal.pone.0015713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JN. Immunity in experimental syphilis. VI Successful vaccination of rabbits with Treponema pallidum, Nichols strain, attenuated by -irradiation. J Immunol. 1973;110:1206–1215. [PubMed] [Google Scholar]
- Mitchell SJ, Engelman J, Kent CK, Lukehart SA, Godornes C, Klausner JD. Azithromycin-resistant syphilis infection: San Francisco, California, 2000-2004. Clin Infect Dis. 2006;42:337–345. doi: 10.1086/498899. [DOI] [PubMed] [Google Scholar]
- Molepo J, Pillay A, Weber B, Morse SA, Hoosen AA. Molecular typing of Treponema pallidum strains from patients with neurosyphilis in Pretoria, South Africa. Sex Transm Infect. 2007;83:189–192. doi: 10.1136/sti.2006.023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monot M, Honoré N, Garnier T, Araoz R, Coppée JY, Lacroix C, Sow S, Spencer JS, Truman RW, Williams DL, Gelber R, Virmond M, Flageul B, Cho SN, Ji B, Paniz-Mondolfi A, Convit J, Young S, Fine PE, Rasolofo V, Brennan PJ, Cole ST. On the origin of leprosy. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- Morgan CA, Lukehart SA, Van Voorhis WC. Immunization with the N-terminal portion of Treponema pallidum repeat protein K attenuates syphilitic lesion development in the rabbit model. Infect Immun. 2002b;70:6811–6816. doi: 10.1128/IAI.70.12.6811-6816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CA, Lukehart SA, Van Voorhis WC. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect Immun. 2003;71:5605–5612. doi: 10.1128/IAI.71.10.5605-5612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CA, Molini BJ, Lukehart SA, Van Voorhis WC. Segregation of B and T cell epitopes of Treponema pallidum repeat protein K to variable and conserved regions during experimental syphilis infection. J Immunol. 2002a;169:952–957. doi: 10.4049/jimmunol.169.2.952. [DOI] [PubMed] [Google Scholar]
- Mulligan CJ, Norris SJ, Lukehart SA. Molecular studies in Treponema pallidum evolution: toward clarity? PloS Negl Trop Dis. 2008;2:e184. doi: 10.1371/journal.pntd.0000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo P. On the American Indian origin of syphilis: fallacies and errors. Allergy Proc. 1994;15:89–99. doi: 10.2500/108854194778703044. [DOI] [PubMed] [Google Scholar]
- Nichols HJ, Hough WH. Demonstration of Spirochaeta pallida in the cerebrospinal fluid. JAMA-J Am Med Assoc. 1913;60:108–110. [Google Scholar]
- Noordhoek GT, Hermans PW, Paul AN, Schouls LM, van der Sluis JJ, van Embden JD. Treponema pallidum subspecies pallidum (Nichols) and Treponema pallidum subspecies pertenue (CDC 2575) differ in at least one nucleotide: comparison of two homologous antigens. Microb Pathog. 1989;6:29–42. doi: 10.1016/0882-4010(89)90005-3. [DOI] [PubMed] [Google Scholar]
- Noordhoek GT, Wieles B, van der Sluis JJ, van Embden JD. Polymerase chain reaction and synthetic DNA probes: a means of distinguishing the causative agents of syphilis and yaws? Infect Immun. 1990;58:2011–2013. doi: 10.1128/iai.58.6.2011-2013.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer GH, Bankhead T, Lukehart SA. ‘Nothing is permanent but change’-antigenic variation in persistent bacterial pathogens. Cell Microbiol. 2009;11:1697–1705. doi: 10.1111/j.1462-5822.2009.01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T, Busch JD, Ravel J, Read TD, Rhoton SD, U'Ren JM, Simonson TS, Kachur SM, Leadem RR, Cardon ML, Van Ert MN, Huynh LY, Fraser CM, Keim P. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci USA. 2004;101:13536–13541. doi: 10.1073/pnas.0403844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay A, Liu H, Chen CY, Holloway B, Sturm AW, Steiner B, Morse SA. Molecular subtyping of Treponema pallidum subspecies pallidum. Sex Transm Dis. 1998;25:408–414. doi: 10.1097/00007435-199809000-00004. [DOI] [PubMed] [Google Scholar]
- Pillay A, Liu H, Ebrahim S, Chen CY, Lai W, Fehler G, Ballard RC, Steiner B, Sturm AW, Morse SA. Molecular typing of Treponema pallidum in South Africa: Cross-Sectional Studies. J Clin Microbiol. 2002;40:256–258. doi: 10.1128/JCM.40.1.256-258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope V, Fox K, Liu H, Marfin AA, Leone P, Seña AC, Chapin J, Fears MB, Markowitz L. Molecular subtyping of Treponema pallidum from North and South Carolina. J Clin Microbiol. 2005;43:3743–3746. doi: 10.1128/JCM.43.8.3743-3746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell ML, Cook DC. The myth of syphilis: The natural history of treponematosis in North America. University Press of Florida, Gainsville: 2005. p. 544. [Google Scholar]
- Rattei T, Ott S, Gutacker M, Rupp J, Maass M, Schreiber S, Solbach W, Wirth T, Gieffers J. Genetic diversity of the obligate intracellular bacterium Chlamydophila pneumoniae by genome-wide analysis of single nucleotide polymorphism: evidence for highly clonal population structure. BMC Genomics. 2007;8:355. doi: 10.1186/1471-2164-8-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román GC, Román L. Occurence of congenital, cardiovascular, visceral, neurologic, and neuro-ophthalmologic complications in late yaws: a theme for future research. Rev Infect Dis. 1986;8:760–777. [PubMed] [Google Scholar]
- Rompalo AM. Can syphilis be eradicated from the world? Curr Opin Infect Dis. 2001;14:41–44. doi: 10.1097/00001432-200102000-00008. [DOI] [PubMed] [Google Scholar]
- Rothschild BM. History of syphilis. Clin Infect Dis. 2005;40:1454–1463. doi: 10.1086/429626. [DOI] [PubMed] [Google Scholar]
- Rothschild C, Rothschild BM. Occurrence and transitions among the treponematoses in North America. Chungara, Revista de Antropologia Chilena. 2000;32:147–155. [Google Scholar]
- Roumagnac P, Weill FX, Dolecek C, Baker S, Brisse S, Chinh NT, Le TAH, Acosta CJ, Farrar J, Dougan G, Achtman M. Evolutionary history of Salmonella typhi. Science. 2006;314:1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders NJ, Jeffries AC, Peden JF, Hood DW, Tettelin H, Rappuoli R, Moxon ER. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol. 2000;37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- Schell RF, Azadegan AA, Nitskansky SG, LeFrock JL. Acquired resistance of hamsters to challenge with homologous and heterologous virulent treponemes. Infect Immun. 1982;37:617–621. doi: 10.1128/iai.37.2.617-621.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay J, Durkin SA, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek J, Ayodeji B, Shatsman S, McLeod MP, Šmajs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Xiang Q, Sodergren E, Baca E, Weinstock GM, Norris SJ, Fraser CM, Paulsen IT. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmajs D, McKevitt M, Howell JK, Norris SJ, Cai WW, Palzkill T, Weinstock GM. Transcriptome of Treponema pallidum: Gene expression profile during experimental rabbit infection. J Bacteriol. 2005;187:1866–1874. doi: 10.1128/JB.187.5.1866-1874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmajs D, McKevitt M, Wang L, Howell JK, Norris SJ, Palzkill T, Weinstock GM. BAC library of T pallidum DNA in E. coli. Genome Res. 2002;12:515–522. doi: 10.1101/gr.207302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmajs D, Zobaníková M, Strouhal M, Čejková D, Dugan-Rocha S, Pospíšilová P, Norris SJ, Albert T, Qin X, Hallsworth-Pepin K, Buhay C, Muzny DM, Chen L, Gibbs RA, Weinstock GM. Complete genome sequence of Treponema paraluiscuniculi, strain Cuniculi A: the loss of infectivity to humans is associated with genome decay. PLoS One. 2011;6:e20415. doi: 10.1371/journal.pone.0020415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert RM. Genus III: Treponema Schaudinn 1905, 1728AL. In: Krieg NR, Holt JG, editors. Bergey's Manual of Systematic Bacteriology. Vol. 1. The Williams & Wilkins Co; Baltimore: 1984. pp. 49–57. [Google Scholar]
- Smith JL. Neuro-ophthalmological study of late yaws. I. An introduction to yaws. Brit J Vener Dis. 1971;47:223–225. doi: 10.1136/sti.47.4.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, David NJ, Indgin S, Israel CW, Levine BM, Justice J, Jr, McCrary JA, 3rd, Medina R, Paez P, Santana E, Sarkar M, Schatz NJ, Spitzer ML, Spitzer WO, Walter EK. Neuro-ophthalmological study of late yaws and pinta II. The Caracas project. Br J Vener Dis. 1971;47:226–251. doi: 10.1136/sti.47.4.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Pesetsky BR. The current status of Treponema cuniculi: Review of the literature. Br J Vener Dis. 1967;43:117–127. doi: 10.1136/sti.43.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Bergen HL. A point mutation associated with bacterial macrolide resistance is present in both 23S rRNA genes of an erythromycin-resistant Treponema pallidum clinical isolate. Antimicrob Agents Chemother. 2000;44:806–807. doi: 10.1128/aac.44.3.806-807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Gherardini FC, Parrish EA, Moomaw CR. Heat shock response of spirochetes. Infect Immun. 1991;59:1572–1575. doi: 10.1128/iai.59.4.1572-1575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Greene SR, Bergen HL, Hardham JM, Barnes NY. Identification and sequence analysis of Treponema pallidum tprJ, a member of a polymorphic multigene family. FEMS Microbiol Lett. 1998;169:155–163. doi: 10.1111/j.1574-6968.1998.tb13312.x. [DOI] [PubMed] [Google Scholar]
- Stamm LV, Kerner TC, Jr, Bankaitis VA, Bassford PJ., Jr Identification and preliminary characterization of Treponema pallidum protein antigens expressed in Escherichia coli. Infect Immun. 1983;41:709–721. doi: 10.1128/iai.41.2.709-721.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouhal M, Šmajs D, Matějková P, Sodergren E, Amin AG, Howell JK, Norris SJ, Weinstock GM. Genome differences between Treponema pallidum subsp. pallidum strain Nichols and T. paraluiscuniculi strain Cuniculi A. Infect Immun. 2007;75:5859–5866. doi: 10.1128/IAI.00709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MY, Liu H, Steiner B, Pillay A, Mickey T, Finelli L, Morse S, Markowitz LE, St Louis ME. Molecular subtyping of Treponema pallidum in an Arizona County with increasing syphilis morbidity: use of specimens from ulcers and blood. J Infect Dis. 2001;183:1601–1606. doi: 10.1086/320698. [DOI] [PubMed] [Google Scholar]
- Tantalo LC, Lukehart SA, Marra CM. Treponema pallidum strain-specific differences in neuroinvasion and clinical phenotype in a rabbit model. J Infect Dis. 2005;191:75–80. doi: 10.1086/426510. [DOI] [PubMed] [Google Scholar]
- Tipple C, McClure MO, Taylor GP. High prevalence of macrolide resistant Treponema pallidum strains in a London centre. Sex Transm Infect. 2011;87:486–488. doi: 10.1136/sextrans-2011-050082. [DOI] [PubMed] [Google Scholar]
- Titz B, Rajagopala SV, Goll J, Häuser R, McKevitt MT, Palzkill T, Uetz P. The binary protein interactome of Treponema pallidum-the syphilis spirochete. PLoS One. 2008;3:e2292. doi: 10.1371/journal.pone.0002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TB, Hollander DH. Biology of the treponematoses based on studies carried out at the International Treponematosis Laboratory Center of the Johns Hopkins University under the auspices of the World Health Organization. Monogr Ser World Health Organ. 1957;35:3–266. [PubMed] [Google Scholar]
- Van Damme K, Behets F, Ravelomanana N, Godornes C, Khan M, Randrianasolo B, Rabenja NL, Lukehart S, Cohen M, Hook E. Evaluation of azithromycin resistance in Treponema pallidum specimens from Madagascar. Sex Transm Dis. 2009;36:775–776. doi: 10.1097/OLQ.0b013e3181bd11dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, Ravel J, Zanecki SR, Pearson T, Simonson TS, U'Ren JM, Kachur SM, Leadem-Dougherty RR, Rhoton SD, Zinser G, Farlow J, Coker PR, Smith KL, Wang B, Kenefic LJ, Fraser-Liggett CM, Wagner DM, Keim P. Global genetic population structure of Bacillus anthracis. PLoS One. 2007;2:e461. doi: 10.1371/journal.pone.0000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser MB, Ellen RP. New insights into the emerging role of oral spirochaetes in periodontal disease. Clin Microbiol Infect. 2011;17:502–512. doi: 10.1111/j.1469-0691.2011.03460.x. [DOI] [PubMed] [Google Scholar]
- Walker EM, Howell JK, You Y, Hoffmaster AR, Heath JD, Weinstock GM, Norris SJ. Physical map of the genome of Treponema pallidum subsp. pallidum (Nichols) J Bacteriol. 1995;177:1797–1804. doi: 10.1128/jb.177.7.1797-1804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM, Hardham JM, McLeod MP, Sodergren EJ, Norris SJ. The genome of Treponema pallidum: new light on the agent of syphilis. FEMS Microbiol Rev. 1998;22:323–332. doi: 10.1111/j.1574-6976.1998.tb00373.x. [DOI] [PubMed] [Google Scholar]
- Weinstock GM, Norris SJ, Sodergren E, Šmajs D. Identification of virulence genes in silico: infectious disease genomics. In: Brogden KA, editor. Virulence mechanisms of bacterial pathogens. third. American Society for Microbiology Press; Washington, DC: 2000a. pp. 251–261. [Google Scholar]
- Weinstock GM, Šmajs D, Hardham J, Norris SJ. From microbial genome sequence to applications. Res Microbiol. 2000b;151:151–158. doi: 10.1016/s0923-2508(00)00115-7. [DOI] [PubMed] [Google Scholar]
- Wendel GD, Jr, Sanchez PJ, Peters MT, Harstad TW, Potter LL, Norgard MV. Identification of Treponema pallidum in amniotic fluid and fetal blood from pregnancies comlicated by congenital syphilis. Obstet Gynecol. 1991;78:890–895. [PubMed] [Google Scholar]
- World Health Organization. The world health report 1998 - Life in the 21st century: A vision for all. Geneva: World Health Organization; 1998. p. 132. [Google Scholar]
- Woznicová V, Matějková P, Flasarová M, Zákoucká H, Vališová Z, Šmajs D, Dastychová E. Clarithromycin treatment failure due to macrolide resistance in Treponema pallidum in a patient with primary syphilis. Acta Derm Venereol. 2010;90:206–207. doi: 10.2340/00015555-0774. [DOI] [PubMed] [Google Scholar]
- Woznicová V, Šmajs D, Wechsler D, Matějková P, Flasarová M. Detection of Treponema pallidum subsp. pallidum from skin lesions, serum, and cerebrospinal fluid in an infant with congenital syphilis after clindamycin treatment of the mother during pregnancy. J Clin Microbiol. 2007;45:659–661. doi: 10.1128/JCM.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]