Abstract
Preeclampsia is a common, pregnancy-specific disorder characterized by reduced placental perfusion, endothelial dysfunction, elevated blood pressure, and proteinuria. The pathogenesis of this heterogeneous disorder is incompletely understood, but it has a familial component, which suggests that one or more common alleles may act as susceptibility genes. We hypothesized that, in a founder population, the genetic background of preeclampsia might also show reduced heterogeneity, and we have performed a genomewide scan in 15 multiplex families recruited predominantly in the Kainuu province in central eastern Finland. We found two loci that exceeded the threshold for significant linkage: chromosome 2p25, near marker D2S168 (nonparametric linkage [NPL] score 3.77; P=.000761) at 21.70 cM, and 9p13, near marker D9S169 (NPL score 3.74; P=.000821) at 38.90 cM. In addition, there was a locus showing suggestive linkage at chromosome 4q32 between D4S413 and D4S3046 (NPL score 3.13; P=.003238) at 163.00 cM. In the present study the susceptibility locus on chromosome 2p25 is clearly different (21.70 cM) from the locus at 2p12 found in an Icelandic study (94.05 cM) and the locus at 2q23 (144.7 cM) found in an Australian/New Zealand study. The locus at 9p13 has been shown to be a candidate region for type 2 diabetes in two recently published genomewide scans from Finland and China. The regions on chromosomes 2p25 and 9p13 may harbor susceptibility genes for preeclampsia.
Preeclampsia is a common (3%) disorder of pregnancy, which, together with thrombosis, is the leading cause of maternal mortality in the Western world. It is a pregnancy-specific vascular disorder characterized by reduced placental perfusion, endothelial dysfunction, and activation of coagulation (Roberts et al. 1989). New-onset hypertension and proteinuria—the symptoms easiest to recognize—develop in the latter half of pregnancy. The progression of the disease is unpredictable, sometimes leading to a convulsive phase, eclampsia. The only effective treatment in eclampsia is the removal of placenta, often leading to premature delivery. The reduced placental perfusion due to abnormal implantation and/or maternal preexisting microvascular disease, such as diabetes and hypertension, is important in the pathophysiology. However, the condition also requires systemic maternal response to the reduced placental perfusion (Ness and Roberts 1996). This response consists of activation of maternal endothelium, accompanied by increased sensitivity of the vasculature to any pressor agent and decreased perfusion to all organs (Roberts and Cooper 2001). Many risk factors of preeclampsia, such as obesity and high blood pressure, increase the risk of atherosclerosis in later life. Indeed, the risk of dying of ischemic heart disease has been shown to be increased in women with a history of preeclampsia (Jonsdottir et al. 1995).
Preeclampsia has a clearly familial component (Chesley and Cooper 1986; Arngrimsson et al. 1990). The incidence of preeclampsia among primigravid women with a family history of preeclampsia is three times that among primigravid women without such family history (Esplin et al. 2001). The central role of the placenta in the disease process may imply that paternal genes present in the fetus contribute to the disease process. If a woman becomes pregnant by a man who has already fathered a preeclamptic pregnancy with a different woman, her risk of developing preeclampsia is increased almost twofold (Lie et al. 1998). Also, a man born of a pregnancy complicated by preeclampsia is twice as likely to father a child from a preeclamptic pregnancy than is a man born of an unaffected pregnancy. On the other hand, infrequent concordance among MZ twins does not support a strong overall genetic effect in this condition (Thornton and Macdonald 1999; Treloar et al. 2001). However, the maternal susceptibility genes are likely to be operative in familial preeclampsia. Identification of those genes will enhance our understanding of the disease mechanism and will make it possible to identify women who are at increased risk for developing preeclampsia.
The genetics of preeclampsia has been difficult to study, because the disease occurs only in women who have reproduced and because its causes are heterogeneous. Candidate gene studies of the angiotensinogen gene, the factor V gene, and the methylenetetrahydrofolate reductase gene on chromosome 1 and the human leukocyte antigen and tumor necrosis factor gene families on chromosome 6 have yielded conflicting results (Broughton Pipkin 1999). However, a whole-genome linkage study provides powerful means to study disease-susceptibility genes. In the first genomewide scan for preeclampsia, a recessive model of inheritance was used for analysis (Hayward et al. 1992). Two whole-genome linkage studies have found a maternal susceptibility locus for preeclampsia on chromosome 2. A significant locus on 2p13 (to D2S286 at 94.05 cM, LOD=4.70) was detected in the Icelandic study that also included patients without proteinuria (Arngrimsson et al. 1999). With similar criteria, a genomewide scan of families from Australia and New Zealand showed a suggestive linkage to 2q (between D2S112 and D2S151 at 144.7 cM, LOD=2.58) (Moses et al. 2000). On the other hand, the genomewide scan of Dutch affected sib-pair families failed to detect linkage to chromosome 2 (Lachmeijer et al. 2001). These three studies were not able to confirm previously suggested linkages to 4q (Harrison et al. 1997) and 7q36 (Arngrimsson et al. 1997; Guo et al. 1997).
We performed a genomewide scan on Finnish families with preeclampsia, with particular emphasis on chromosomes 1, 6, and 2p13. We hypothesized that the genetic mechanisms underlying preeclampsia might have reduced heterogeneity, because the genetic background of the Finnish founder subpopulations might be more uniform than those of outbred populations (de la Chapelle and Wright 1998; Peltonen et al. 1999, 2000; Kere 2001; Laitinen et al. 2001).
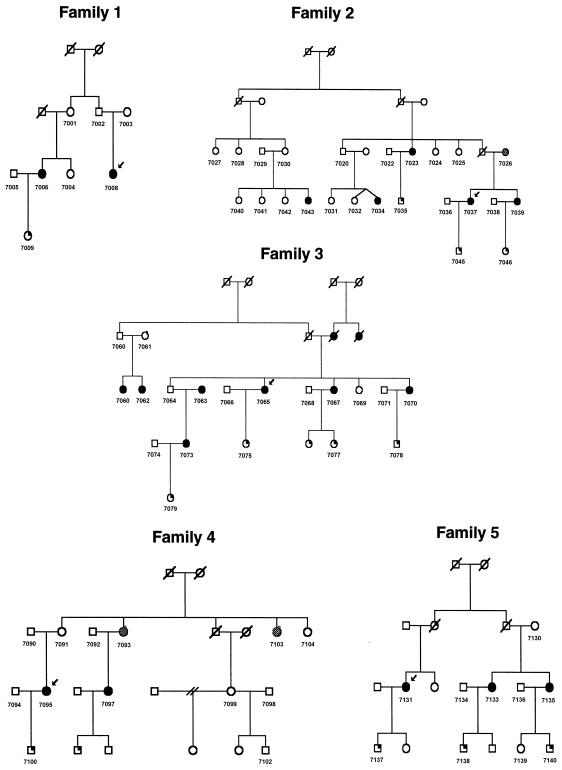
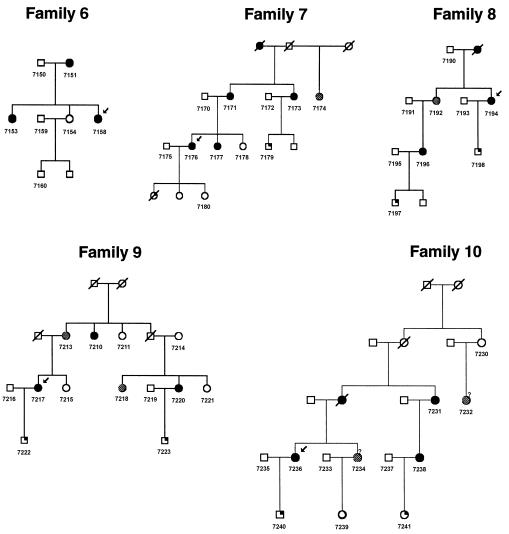
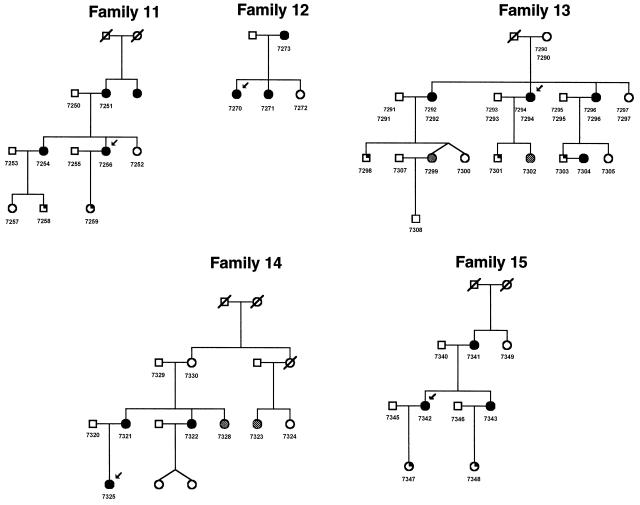
Families with preeclampsia were recruited predominantly from the Kainuu province in central eastern Finland, the location of one of the rural populations that resulted from the internal migration in the 16th century, and in the Helsinki region in southern Finland. With the approval of the local ethical committees, we recruited the preeclampsia probands by sending letters to the patients who had been hospitalized with the diagnosis of severe preeclampsia (see definition below) in their first pregnancies either in Kainuu Central Hospital, Kajaani, or the Department of Obstetrics and Gynecology, Helsinki University Central Hospital, Helsinki, between 1988 and 1997, and by advertisements for the study. Between June 1997 and April 1999, we recruited 15 multiplex families (174 individuals) through the probands. Pedigrees 1–10 (figs. 1 and 2) were recruited from the Kainuu region, whereas pedigrees 11–15 were recruited from Helsinki (fig. 3). To verify the origin of families, we traced the birthplaces of parents and grandparents, using church records. All participants gave their informed written consent. The reproductive medical records of all women were reviewed to confirm or exclude the diagnosis. Forty-eight women met the criteria of preeclampsia in their first pregnancies and one woman in her second pregnancy. One woman met the criteria of eclampsia in her first pregnancy. Three families had two affected individuals, eight families had three affected, two families had four affected, one family had five affected and one family had seven affected. Severe preeclampsia was diagnosed in 29 of these 50 women (58%), including all probands. Thirteen women met the criteria of gestational hypertension in their first pregnancies. Every family was ascertained on the basis of at least one proteinuric preeclampsia case. Within ascertained families, women who had gestational hypertension without proteinuria were also considered as affected under the general criteria, because of the higher likelihood of shared susceptibility genes within families. Preeclampsia was defined as systolic blood pressure >140 mm Hg or diastolic blood pressure >90 mm Hg in a woman who was normotensive before 20 wk of gestation, and the urinary excretion of ⩾0.3 g protein in a 24-h specimen or ⩾1+ reading on dipstick in a random urine determination at least twice with no evidence of the urinary tract infection. Severe preeclampsia was defined if one of the following criteria was met: systolic blood pressure ⩾160 mm Hg, diastolic blood pressure ⩾110 mm Hg, or proteinuria ⩾2.0 g in 24 h. Eclampsia was defined as the occurrence, in a woman with preeclampsia, of seizures that could not be attributed to other causes. Hypertension and proteinuria were confirmed to have disappeared 3 mo after delivery. In families that included at least one patient with preeclampsia or severe preeclampsia, women with gestational hypertension (systolic blood pressure >140 mm Hg or diastolic blood pressure >90 mm Hg in a woman who was normotensive before 20 wk of gestation) without significant proteinuria (<0.3 g/24 h) were considered to have mild preeclampsia. As in genomewide scan studies published elsewhere, we used two diagnostic schemes in the present study. According to the strict criteria, we included only preeclamptic and eclamptic women, and, according to the general criteria, we also included women with mild preeclampsia (gestational hypertension). Of the affected women, 79% (50/63) met the strict criteria, and 21% (13/63) met only the general criteria. Five families (families 4, 9, 10, 13, and 14) contained two women with a history of gestational hypertension, and three families (families 2, 7, and 8) contained one woman such a history (figs. 1–3).
Figure 1.
Pedigrees 1–5 recruited for linkage study from the Kainuu region. Blackened circles indicate preeclamptic and eclamptic women. Shaded circles indicate women with mild preeclampsia (gestational hypertension). Symbols with a blackened upper right quadrant indicate children born from preeclamptic pregnancies.
Figure 2.
Pedigrees 6–10 recruited for linkage study from the Kainuu region. Blackened circles indicate preeclamptic and eclamptic women. Shaded circles indicate women with mild preeclampsia (gestational hypertension). Symbols with a blackened upper right quadrant indicate children born from preeclamptic pregnancies.
Figure 3.
Pedigrees 11–15 recruited for linkage study from Helsinki. Blackened circles indicate preeclamptic and eclamptic women. Shaded circles indicate women with mild preeclampsia (gestational hypertension). Symbols with a blackened upper right quadrant indicate children born from preeclamptic pregnancies.
EDTA-treated blood (40 ml) was collected from each individual. DNA was extracted by a standard nonenzymatic method. For the first stage of analysis, genotyping was performed in the Finnish Genome Center by use of 381 polymorphic microsatellite markers from ABI Prism Linkage Mapping Set–MD10 (Applied Biosystems). The average interval between the markers was 10 cM. DNA (20 ng) was dried on 384-well plates at 37°C overnight before PCR setup with a Tecan Genesis 150 pipetting robot (Biomatic Technologies). The PCRs were performed in 5-μl volumes, using reagent concentrations and temperature profiles as recommended by the reagent manufacturer (Applied Biosystems). The fluorescencelabeled PCR products were pooled into 28 panels according to the Linkage Mapping Set–MD10. The pooled samples were dialyzed before electrophoresis, and electrophoresis was performed on a MegaBace 1,000-capillary electrophoresis instrument (Molecular Dynamics). The results were analyzed and alleles were called using Genetic Profiler v. 1.1 software (Molecular Dynamics). For the second stage of analysis, we designed 21 additional markers (table 1) between original genome scan markers, using the Marshfield comprehensive human genetic map. The PCRs were performed as described above. For the final analysis, we also included markers genotyped in the Department of Medical Genetics, University of Helsinki (20 markers on chromosome 1, 15 markers on chromosome 6 [derived from Weber set 6], and 8 markers on chromosome 2, around marker D2S286, which had the highest nonparametric linkage [NPL] score in the Icelandic study), which were designed using the Marshfield comprehensive human genetic map and clone NHO355F16 (GenBank AC007681). For these additional markers, PCR was performed in 15-μl reaction volumes that contained 50 ng of genomic DNA, together with fluorescently labeled primers. The amplified PCR products were separated using 4.25% polyacrylamide gels run on an ABI 377 sequencer (Applied Biosystems). Lane tracking and allele calling were performed using GeneScan and Genotyper software (Applied Biosystems). Two investigators scored all the gels.
Table 1.
Additional Markers on Chromosomes 2, 4, 9, and 15
| Chromosomeand Marker | BetweenMarkers | HeterozygoteFrequency |
| 2: | ||
| D2S398 | D2S162, D2S168 | .74 |
| D2S2199 | D2S168, D2S305 | .63 |
| D2S149 | D2S168, D2S305 | .72 |
| D2S220 | D2S305, D2S2247 (D2S165) | .77 |
| D2S2247 | Replacing D2S165 | .72 |
| D2S352 | D2S2247 (D2S165), D2S367 | .82 |
| 4: | ||
| D4S1579 | D4S1575, D4S424 | .63 |
| D4S2962 | D4S424, D4S413 | .82 |
| D4S3046 | D4S413, D4S1597 | .77 |
| D4S1502 | D4S1597, D4S1539 | .65 |
| 9: | ||
| D9S289 | D9S1677, D9S1776 | .75 |
| D9S1811 | D9S1776, D9S1682 | .82 |
| D9S1829 | D9S1682, D9S290 | .77 |
| D9S1863 | D9S290, D9S164 | .73 |
| D9S169 | D9S171, D9S161 | .83 |
| D9S319 | D9S161, D9S1817 | .80 |
| D9S1874 | D9S1817, D9S273 | .84 |
| 15: | ||
| D15S1048 | D15S1002, D15S165 | .84 |
| D15S1010 | D15S165, D15S1007 | .80 |
| D15S1040 | D15S1007, D15S1012 | .77 |
| GAAC1C11 | D15S1007, D15S1012 | .87 |
The total scan included 435 microsatellite markers. The data were Mendel checked with Pedmanager and Pedcheck softwares, and were haplotyped with Genehunter 2.1 software (Whitehead Institute for Biomedical Research), to check for double recombinants. Linkage was assessed by the affecteds-only multipoint NPL analysis method of Genehunter v. 2.1, because the mode of inheritance of preeclampsia is unknown. Women who were not known to be affected or were not confirmed to have had normal pregnancies or who had not reproduced—and all men—were classified as “unknown.” The threshold for genomewide significant linkage was determined by simulation according to the principles set forth by Lander and Kruglyak (1995).
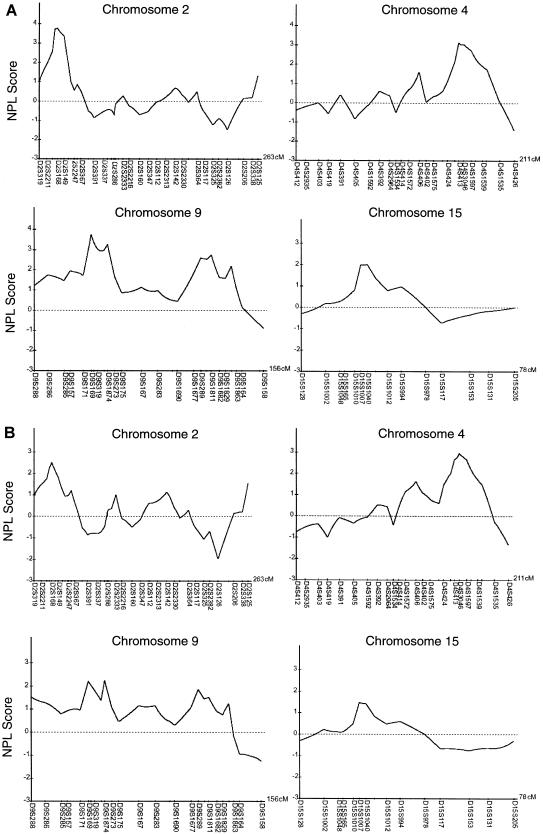
The initial genome scan on 381 markers revealed five loci when strict criteria were used and six loci when general criteria were used, thus exceeding an NPL score of 2. We chose five regions for further mapping with a denser set of markers (table 1). We found a significant linkage to chromosome 2p25 near marker D2S168 (NPL score 3.77; P=.000761) at 21.70 cM and to chromosome 9p13 near marker D9S169 (NPL score 3.74; P=.000821) at 38.90 cM, and we found suggestive linkage to chromosome 4q32 between D4S413 and D4S3046 (NPL score 3.13; P=.003238) at 163.00 cM and chromosome 9p11 near marker D9S1874 (NPL score 3.26; P=.002456) at 49.90 cM when the general criteria were used (fig. 4A). Moreover, the following regions displayed nominal linkage: chromosome 9q33 near marker D9S1811 (NPL score 2.74; P=.007436) at 120.80 cM, 9q34 near marker D9S1863 (NPL score 2.18; P=.022627) at 134.70 cM, and 15q11 between markers D15S1007 and D15S1040 (NPL score 2.00; P=.031434) at 24.50 cM.
Figure 4.
Multipoint NPL analysis results, from chromosomes 2, 4, 9, and 15, for the preeclampsia genomewide scan. A, Results when the general criteria were used. B, Results when the strict criteria were used. The Y-axis indicates the NPL score. The X-axis indicates the markers and the length of the chromosome.
When the strict criteria were used, the NPL scores were 2.51 for chromosome 2p25 (P=.009919), 2.22 for 9p13 (P=.018194), and 2.96 for 4q32 (P=.018194) (fig. 4B). In addition, chromosome 9p11 displayed nominal linkage (NPL score 2.20; P=.019089).
The loci 2p25 and 9p13 correlated in five families (families 6, 8, 10, 11, and 13) when general criteria were used and in four families (families 6, 10, 11, and 13) when strict criteria were used (figs. 1–3). The loci 9p13 and 4q32 correlated in six families (families 1, 2, 6, 11, 12, and 13 when either general or strict criteria were used (figs. 1, 2, and 3). The loci 2p25 and 4q32 correlated in seven families (families 5, 6, 7, 11, 13, 14, and 15) when general criteria were used and in eight families (families 5, 6, 7, 10, 11, 13, 14, and 15) when strict criteria were used (figs. 1, 2, and 3).
The correspondence of the most significant loci in each family was very good when the results of the two criteria were compared. The NPL scores for the two most significant loci, 2p25 and 9p13, were reduced almost equally (1.26–1.52) when the strict criteria were used and number of affecteds was reduced from 63 to 50. Interestingly, the NPL level for the 4q32 locus was reduced by only 0.17 NPL scores, and thus there may be differences between the effect of the different loci.
In the present genomewide scan for preeclampsia, we studied 15 families collected, in part, from a founder subpopulation in Finland, which may offer advantages for the mapping of complex genetic traits (de la Chapelle and Wright 1998; Peltonen et al. 1999, 2000; Kere 2001; Laitinen et al. 2001). For maximizing genetic homogeneity, we recruited these families predominantly from Kainuu province in central eastern Finland. As was done in two other genomewide scans for preeclampsia, one of subjects from Iceland (Arngrimsson et al. 1999) and one of subjects from Australia and New Zealand (Moses et al. 2000), we used both the general criteria, which included women with prior preeclampsia, eclampsia, or gestational hypertension, and the strict criteria, which included women with prior preeclampsia or eclampsia.
We found a significant linkage to chromosome 2p25. In two recent genomewide scans, the most interesting regions have also been found on chromosome 2. In the Icelandic study, the support for the locus on 2p13, with a LOD score of 4.70, came mostly from two large families, and the most telomeric 2p marker compatible with linkage in at least one of the families was D2S2368 at position 89.24 cM in the most recent deCODE linkage map (Arngrimsson et al. 1999; Kong et al. 2002). In the Australian/New Zealand study, the most interesting region was on 2q23, with a LOD score of 2.58; the peak is telomeric to D2S112 at 145.71 cM in the deCODE map (Moses et al. 2000). Although the distance between these two loci is ∼50 cM, the investigators concluded that it is likely that both studies have detected the same locus, and the locus has been designated “PEE1,” for preeclampsia, eclampsia gene-1. We now present evidence for a new locus on 2p that by genetic distance is unlinked to the previous loci. The 95% CI for our linkage result already excludes D2S2247 at 50.34 cM (deCODE map distance), with a peak toward the telomere of 2p. With an almost 40-cM gap between the confidence limits for the Icelandic and Finnish mapping results, and nearly 100 cM between the Finnish and the Australian/New Zealand peak, it is inconceivable that the three linkage results would detect one and the same locus. Thus, both the number and locations of preeclampsia-susceptibility gene(s) on chromosome 2 remain ambiguous. There are previous examples of the unexpected mapping of more than one locus on the same chromosome for a single phenotype, but the nature of the genes is not known in any of those. It is possible that members of the same gene family, with partially redundant and overlapping effects, might account for such mapping results; indeed, segmental duplications are prominent at the centromere and on the long arm of chromosome 2 (Bailey et al. 2002). Alternatively, genes of the same regulatory pathway may cause highly similar phenotypes with different map positions (e.g., Kere and Elomaa 2002).
The second significant finding came from chromosome 9p13. Interestingly, it has been shown to be a candidate region for type 2 diabetes mellitus in two genomewide scans from the Botnia region in Finland (Lindgren et al. 2002) and China (Luo et al. 2001). Although no hypertensive phenotype of insulin-resistance syndrome has been attributed to chromosome 9, other features of this syndrome (type 2 diabetes mellitus and high-density lipoprotein cholesterol concentrations) have been linked to chromosome 9 (Arya et al. 2002; Lindgren et al. 2002). Phenotype can also change during pregnancy, because pregnancy itself is a highly insulin-resistant state (Kaaja et al. 1999).
The suggestive linkage to chromosome 4q32 in the present study is within the same region that was identified in the earlier of the two Australian genomewide scans, which used both parametric and nonparametric analyses (Harrison et al. 1997). Families in that study are a subset of the authors' later genomewide scan (Moses et al. 2000). Curiously, this finding could not be replicated in the authors' later study with a larger family panel that also included families from New Zealand. It is possible that increased genetic heterogeneity affected the result.
We were not able to detect linkage to chromosome 1, which contains some candidate genes for preeclampsia and demonstrates inconclusive findings such as the angiotensinogen gene (Arngrimsson et al. 1993; Ward et al. 1993; Guo et al. 1997), the factor V gene (O'Shaughnessy et al. 1999; Watanabe et al. 2001) and the methylenetetrahydrofolate reductase gene (Sohda et al. 1997; Powers et al. 1999; Laivuori et al. 2000). As with our study of chromosome 1, we used a higher marker density to study chromosome 6, which contains the human leukocyte antigen and tumor necrosis factor gene families, but we did not find any linkage. Also, endothelial nitric oxide synthase on chromosome 7 has been proposed as a strong candidate gene in pregnancy-induced hypertension (Arngrimsson et al. 1997). The results of the present study gave no support to this finding in the Finnish population. Furthermore, the most recently reported genome scan for preeclampsia of Dutch affected sib pairs did not find any peaks in these regions (Lachmeijer et al. 2001).
Our results provide evidence for linkage of preeclampsia to chromosome 2p25 and 9p13. Our surprising finding of a separate locus for preeclampsia on chromosome 2p25, distinct from two loci reported in two other genome scans, requires further investigation.
Acknowledgments
Above all, we are grateful to all the families who made this study possible. We would like to express our thanks to Ms. Marjatta Vallas, Ms. Leena Järvinen, Ms. Siv Knaappila, Ms. Riitta Lehtinen, and all the personnel of the Finnish Genome Center, for their skillful work in this project, as well as Ms. Janet Lawler-Heavner, for her expertise in the revision of the language of the manuscript. This study was supported by grants from the Academy of Finland, the Biomedicum Foundation, the Finnish Medical Society Duodecim, the Helsingin Sanomat Centennial Foundation, the Jalmari and Rauha Ahokas Foundation, the Maud Kuistila Foundation, and the research funds from the Helsinki University Central Hospital.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation http://research.marshfieldclinic.org/genetics/ (for the additional markers used in the study)
- Entrez Nucleotides Database, http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=Nucleotide (for the clone NHO355F16 [GenBank accession number AC007681])
- University of Helsinki–Finnish Genome Center, http://www.genome.Helsinki.fi/eng/research/projects/pre-eclampsia/markers.txt (for all markers and the genetic distances used in the present study)
- http://lpg.nci.nih.gov/ (for markers derived from Weber set 6)
References
- Arngrimsson R, Bjornsson S, Geirsson RT, Bjornsson H, Walker JJ, Snaedal G (1990) Genetic and familial predisposition to eclampsia and pre-eclampsia in a defined population. Br J Obstet Gynaecol 97:762–769 [DOI] [PubMed] [Google Scholar]
- Arngrimsson R, Hayward C, Nadaud S, Baldursdottir A, Walker JJ, Liston WA, Bjarnadottir RI, Brock DJ, Geirsson RT, Connor JM, Soubrier F, Broughton Pipkin F (1997) Evidence for a familial pregnancy-induced hypertension locus in the eNOS-gene region. Am J Hum Genet 61:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 [DOI] [PubMed] [Google Scholar]
- Arngrimsson R, Purandare S, Connor M, Walker JJ, Bjornsson S, Soubrier F, Kotelevtsev YV, Geirsson RT, Bjornsson H (1993) Angiotensinogen: a candidate gene involved in preeclampsia? Nat Genet 4:114–115 [DOI] [PubMed] [Google Scholar]
- Arngrimsson R, Sigurardottir S, Frigge ML, Bjarnadottir RI, Jonsson T, Stefansson H, Baldursdottir A, Einarsdottir AS, Palsson B, Snorradottir S, Lachmeijer AM, Nicolae D, Kong A, Bragason BT, Gulcher JR, Geirsson RT, Stefansson K (1999) A genome-wide scan reveals a maternal susceptibility locus for pre-eclampsia on chromosome 2p13. Hum Mol Genet 8:1799–1805 [DOI] [PubMed] [Google Scholar]
- Arya R, Duggirala R, Almasy L, Rainwater DL, Mahaney MC, Cole S, Dyer TD, Williams K, Leach RJ, Hixson JE, MacCluer JW, O'Connell P, Stern MP, Blangero J (2002) Linkage of high-density lipoprotein–cholesterol concentrations to a locus on chromosome 9p in Mexican Americans. Nat Genet 30:102–105 [DOI] [PubMed] [Google Scholar]
- Broughton Pipkin F (1999) What is the place of genetics in the pathogenesis of pre-eclampsia? Biol Neonate 76:325–330 [DOI] [PubMed] [Google Scholar]
- Chesley LC, Cooper DW (1986) Genetics of hypertension in pregnancy: possible single gene control of pre-eclampsia and eclampsia in the descendants of eclamptic women. Br J Obstet Gynaecol 93:898–908 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Wright FA (1998) Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA 95:12416–12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplin MS, Fausett MB, Fraser A, Kerber R, Mineau G, Carrillo J, Varner MW (2001) Paternal and maternal components of the predisposition to preeclampsia. N Engl J Med 344:867–872 [DOI] [PubMed] [Google Scholar]
- Guo G, Wilton AN, Fu Y, Qiu H, Brennecke SP, Cooper DW (1997) Angiotensinogen gene variation in a population case-control study of preeclampsia/eclampsia in Australians and Chinese. Electrophoresis 18:1646–1649 [DOI] [PubMed] [Google Scholar]
- Harrison GA, Humphrey KE, Jones N, Badenhop R, Guo G, Elakis G, Kaye JA, Turner RJ, Grehan M, Wilton AN, Brennecke SP, Cooper DW, Broughton Pipkin F (1997) A genomewide linkage study of preeclampsia/eclampsia reveals evidence for a candidate region on 4q. Am J Hum Genet 60:1158–1167 [PMC free article] [PubMed] [Google Scholar]
- Hayward C, Livingstone J, Holloway S, Liston WA, Brock DJ (1992) An exclusion map for pre-eclampsia: assuming autosomal recessive inheritance. Am J Hum Genet 50:749–757 [PMC free article] [PubMed] [Google Scholar]
- Jonsdottir LS, Arngrimsson R, Geirsson RT, Sigvaldason H, Sigfusson N (1995) Death rates from ischemic heart disease in women with a history of hypertension in pregnancy. Acta Obstet Gynecol Scand 74:772–776 [DOI] [PubMed] [Google Scholar]
- Kaaja R, Laivuori H, Laakso M, Tikkanen MJ, Ylikorkala O (1999) Evidence of a state of increased insulin resistance in preeclampsia. Metab Clin Exp 48:892–896 [DOI] [PubMed] [Google Scholar]
- Kere J (2001) Human population genetics: lessons from Finland. Annu Rev Genomics Hum Genet 2:103–128 [DOI] [PubMed] [Google Scholar]
- Kere J, Elomaa O (2002) Healing a natural knockout of epithelial organogenesis. Trends Mol Med 8:197–200 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjatsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallback B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lachmeijer AM, Arngrimsson R, Bastiaans EJ, Frigge ML, Pals G, Sigurdardottir S, Stefansson H, Palsson B, Nicolae D, Kong A, Aarnoudse JG, Gulcher JR, Dekker GA, ten Kate LP, Stefansson K (2001) A genome-wide scan for preeclampsia in the Netherlands. Eur J Hum Genet 9:758–764 [DOI] [PubMed] [Google Scholar]
- Laitinen T, Daly MJ, Rioux JD, Kauppi P, Laprise C, Petäys T, Green T, Cargill M, Haahtela T, Lander ES, Laitinen LA, Hudson TJ, Kere J (2001) A susceptibility locus for asthma-related traits on chromosome 7 revealed by genome-wide scan in a founder population. Nat Genet 28:87–91 [DOI] [PubMed] [Google Scholar]
- Laivuori H, Kaaja R, Ylikorkala O, Hiltunen T, Kontula K (2000) 677 C→T polymorphism of the methylenetetrahydrofolate reductase gene and preeclampsia. Obstet Gynecol 96:277–280 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lie RT, Rasmussen S, Brunborg H, Gjessing HK, Lie-Nielsen E, Irgens LM (1998) Fetal and maternal contributions to risk of pre-eclampsia: population based study. BMJ 316:1343–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren CM, Mahtani MM, Widen E, McCarthy MI, Daly MJ, Kirby A, Reeve MP, Kruglyak L, Parker A, Meyer J, Almgren P, Lehto M, Kanninen T, Tuomi T, Groop LC, Lander ES (2002) Genomewide search for type 2 diabetes mellitus susceptibility loci in Finnish families: the Botnia study. Am J Hum Genet 70:509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo TH, Zhao Y, Li G, Yuan WT, Zhao JJ, Chen JL, Huang W, Luo M (2001) A genome-wide search for type II diabetes susceptibility genes in Chinese Hans. Diabetologia 44:501–506 [DOI] [PubMed] [Google Scholar]
- Moses EK, Lade JA, Guo G, Wilton AN, Grehan M, Freed K, Borg A, Terwilliger JD, North R, Cooper DW, Brennecke SP (2000) A genome scan in families from Australia and New Zealand confirms the presence of a maternal susceptibility locus for pre-eclampsia, on chromosome 2. Am J Hum Genet 67:1581–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness RB, Roberts JM (1996) Heterogeneous causes constituting the single syndrome of preeclampsia: a hypothesis and its implications. Am J Obstet Gynecol 175:1365–1370 [DOI] [PubMed] [Google Scholar]
- O'Shaughnessy KM, Fu B, Ferraro F, Lewis I, Downing S, Morris NH (1999) Factor V Leiden and thermolabile methylenetetrahydrofolate reductase gene variants in an East Anglian preeclampsia cohort. Hypertension 33:1338–1341 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T, Macdonald AM (1999) Molecular genetics of the Finnish disease heritage. Hum Mol Genet 8:1913–1923 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Powers RW, Minich LA, Lykins DL, Ness RB, Crombleholme WR, Roberts JM (1999) Methylenetetrahydrofolate reductase polymorphism, folate, and susceptibility to preeclampsia. J Soc Gynecol Investig 6:74–79 [DOI] [PubMed] [Google Scholar]
- Roberts JM, Cooper DW (2001) Pathogenesis and genetics of pre-eclampsia. Lancet 357:53–56 [DOI] [PubMed] [Google Scholar]
- Roberts JM, Taylor RN, Musci TJ, Rodgers GM, Hubel CA, McLaughlin MK (1989) Preeclampsia: an endothelial cell disorder. Am J Obstet Gynecol 161:1200–1204 [DOI] [PubMed] [Google Scholar]
- Sohda S, Arinami T, Hamada H, Yamada N, Hamaguchi H, Kubo T, Morris NH (1997) Methylenetetrahydrofolate reductase polymorphism and pre-eclampsia. J Med Genet 34:525–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JG, Macdonald AM (1999) Twin mothers, pregnancy hypertension and pre-eclampsia. Br J Obstet Gynaecol 106:570–575 [DOI] [PubMed] [Google Scholar]
- Treloar SA, Cooper DW, Brennecke SP, Grehan MMR, Martin NG (2001) An Australian twin study of the genetic basis of preeclampsia and eclampsia. Am J Obstet Gynecol 184:374–381 [DOI] [PubMed] [Google Scholar]
- Ward K, Hata A, Jeunemaitre X, Helin C, Nelson L, Namikawa C, Farrington PF, Ogasawara M, Suzumori K, Tomoda S, Berrebi S, Sasaki M, Corvol P, Lifton RP, Lalouel JM (1993) A molecular variant of angiotensinogen associated with preeclampsia. Nat Genet 4:59–61 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Hamada H, Yamakawa-Kobayashi K, Yoshikawa H, Arinami T, Guo G, Wilton AN, Fu Y, Qiu H, Brennecke SP, Cooper DW (2001) Evidence for an association of the R485K polymorphism in the coagulation factor V gene with severe preeclampsia from screening 35 polymorphisms in 27 candidate genes. Thromb Haemost 86:1594–1595 [PubMed] [Google Scholar]