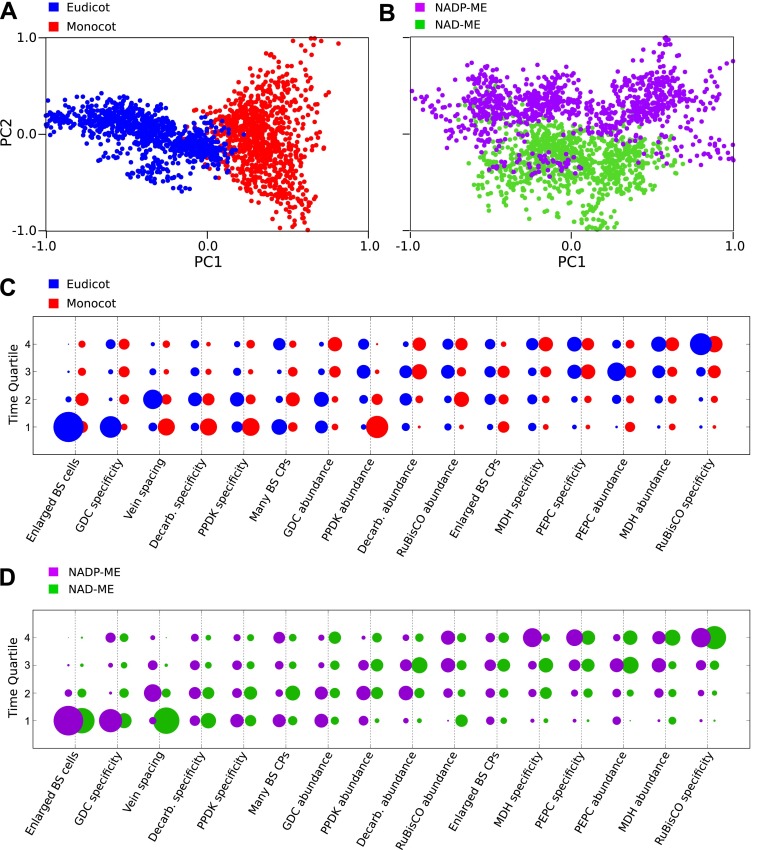
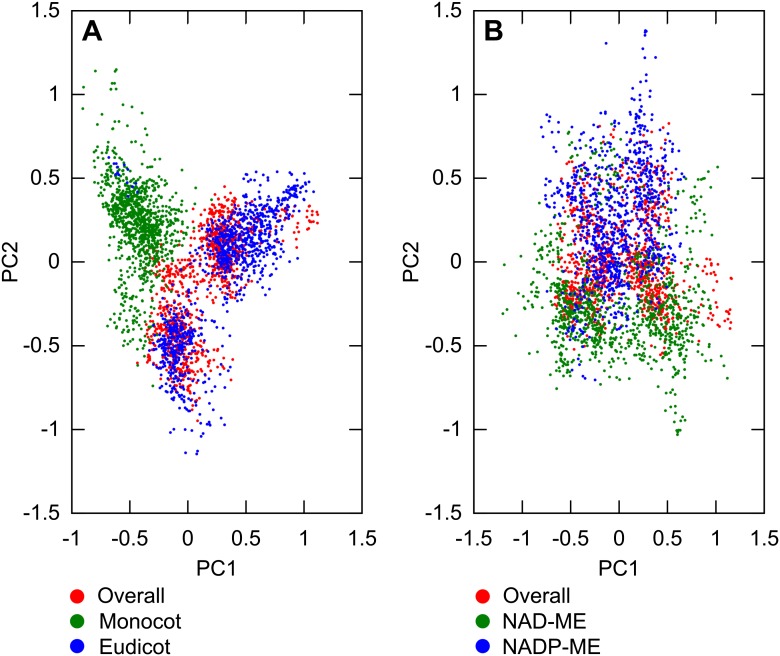
Figure 4. Differences in the evolutionary events generating different C4 sub-types and distantly related taxa.
Principal component analysis (PCA) on the entire landscape of transition probabilities using only monocot and eudicot data (A) and data from NADP-ME and NAD-ME sub-type lineages (B) shows broad differences between the evolutionary pathways generating C4 in each taxon. Monocots and eudicots differ in the predicted timing of events generating C4 anatomy and biochemistry (C), whereas NADP-ME and NAD-ME lineages differ primarily in the evolution of decreased vein spacing and greater numbers of chloroplasts in BS cells (D).