Figure 6.
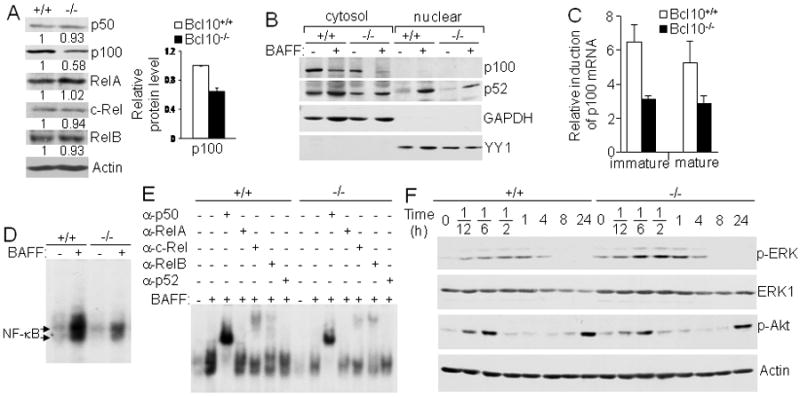
Bcl10-deficient splenic B cells express reduced levels of p100, indirectly affecting BAFF-induced noncanonical NF-κB activation. A, Expression of NF-κB/Rel family members. Protein levels of NF-κB/Rel family members in total cellular extracts from splenic mature B cells were determined by direct Western blotting analysis with the indicated antibodies. Densitometric analysis of protein bands was performed and normalized to actin loading control. Levels of each NF-κB protein in Bcl10-deficient and wild-type B cells were compared by giving wild-type an arbitrary value of 1. The bar graph shows the relative p100 protein levels calculated from 3 independent experiments. p < 0.01. B, BAFF-induced NF-κB2/p100 processing and nuclear translocation. Following 16 hours of culture of splenic mature B cells with or without BAFF, cytoplasmic and nuclear extracts were prepared and subjected to Western blotting with the indicated antibodies. GAPDH was used as cytoplasmic protein loading control and YY1 as nuclear protein loading control. C, BAFF-induced NF-κB2/p100 mRNA expression. Splenic immature (CD93+) and mature (CD93-CD23+) B cells were either stimulated with BAFF or left non-stimulated for 16 hrs. NF-κB2/p100 mRNA expression was determined by quantitative real-time PCR. Relative fold induction in response to BAFF was normalized to 18S rRNA and calibrated to non-stimulated sample for the corresponding genotype. D and E, BAFF-induced noncanonical activation of NF-κB. Splenic mature B cells were constantly stimulated with BAFF for 16 hrs and total cell lysates were subjected to NF-κB gel mobility shift (D) and antibody supershift (E) analysis. Arrows indicate the position of NF-κB complexes. F, BAFF-induced ERK and Akt activation. Splenic mature B cells were stimulated with BAFF for the indicated times and cell lysates were subjected to direct Western blotting analysis with the indicated antibodies. Data are representative of 3 (A, B, D and E) or 2 (F) independent experiments, or data is representative of at least 3 experiments that used one animal from each genotype per experiment (C).
