Table 1. Partial RMSDs for the MD-NMR simulation.
| Segment | Definition | RMSD(Å) |
| H1 | 11–18 | 7.3 |
| H2 | 24–34 | 3.5 |
| H3 | 45–54 | 2.8 |
| H4 | 62–72 | 2.6 |
| H5 | 82–93 | 1.6 |
| H6 | 98–108 | 1.8 |
| H7 | 118–132 | 1.3 |
| H8 | 146–155 | 5.0 |
| H9 | 166–174 | 5.0 |
| β1 | 42–44 | 1.9 |
| β2 | 58–60 | 3.1 |
| β3 | 79–81 | 1.8 |
| β4 | 115–117 | 2.0 |
| β5 | 136–138 | 11.6 |
| β6 | 163–165 | 3.3 |
| L1 | 56–61 | 3.7 |
| L2 | 133–145 | 10.2 |
| L3 | 175–187 | 8.3 |
Partial RMSD evaluated after structure alignment over the backbone atoms of protein core residues. RMSD values for the helices H1 to H9, the 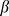 -strands
-strands 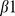 to
to 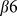 , and the loops L1, L2, and L3 between the most representative structure of the MD-NMR trajectory and the corresponding initial structure.
, and the loops L1, L2, and L3 between the most representative structure of the MD-NMR trajectory and the corresponding initial structure. 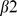 and
and 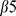 are sub-structures of the L1 and L2 segments respectively.
are sub-structures of the L1 and L2 segments respectively.
