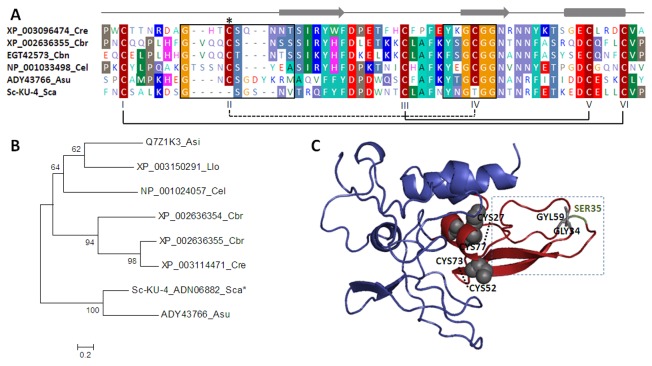
Figure 1. Sequence, phylogeny and structural analysis of Sc-KU-4.
A: Amino acid sequence alignment of the Kunitz domain in Sc-KU-4 with the homologous domains of other nematode Kunitz-like inhibitors. Conserved positions are shaded; the solid lines indicate the conserved disulfide bridges, and the discontinuous line indicates the missing CysII–CysIV bridge in Sc-KU-4. B: Phylogenetic analysis showing Sc-KU-4 closes to other nematode Kunitz-like inhibitors forming a separate cluster with A. suum. C: Sc-KU-4 structure prediction based on multiple-threading alignments showing the Kunitz domain exposed on the molecular surface in a characteristic tertiary fold of three antiparallel β-strands followed by one α-helix. Cysteine residues responsible for reactive loop stabilization and glycine residues that replace CysII–CysIV are shown in gray. The discontinuous line box highlights the amino acids present in reactive loops. The P1 position is shown in green. Asi: Anisakis simplex; Asu: Ascaris suum; Cbn: Caenorhabditis brenneri; Cbr: C. briggsae; Cel: C. elegans; Cre: C. remanei; Llo: Loa loa; Sca: S. carpocapsae.