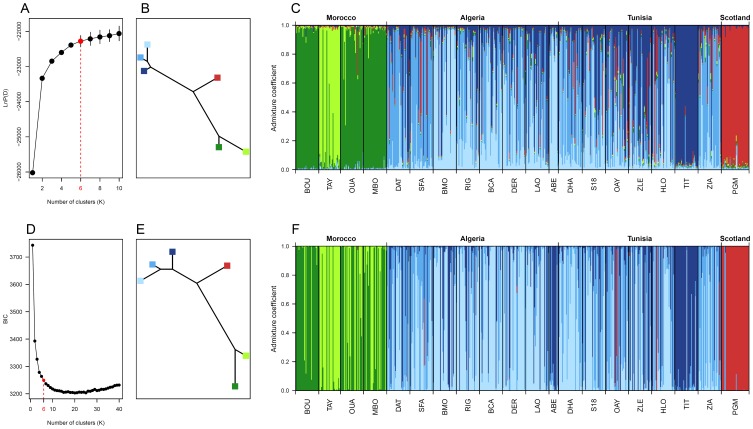
Figure 2. Regional genetic structure of A. glutinosa as inferred by Structure (A, B and C) and DAPC (D, E and F).
Results from the population genetics model-based Bayesian clustering method implemented in Structure: (A) variation of the likelihood of the data across a range of number of clusters K; (B) neighbour joining tree computed using the genetic distance (allele frequency divergence) between clusters; (C) histogram of individual assignment to clusters where each individual is represented by a thin vertical bar partitioned into several coloured segments proportionally to its membership of a given cluster (admixture coefficient). Results from the multivariate statistics based clustering method implemented in DAPC: (D) variation of the Bayesian Information Criterion (BIC) of the k-means clustering algorithm across a range of number of clusters K; (E) neighbour joining tree computed using the DAPC distance between centroids of the clusters; (F) histogram of individual assignment to clusters where each individual is represented by a thin vertical bar partitioned into several coloured segments proportionally to its membership of a given cluster (admixture coefficient). All graphs were plotted in R v.2.12.0 [36].