Abstract
AIM: To investigate the effect of retinoblastoma protein-interacting zinc finger gene 1 (RIZ1) upregulation in gene expression profile and oncogenicity of human esophageal squamous cell carcinoma (ESCC) cell line TE13.
METHODS: TE13 cells were transfected with pcDNA3.1(+)/RIZ1 and pcDNA3.1(+). Changes in gene expression profile were screened and the microarray results were confirmed by reverse transcription-polymerase chain reaction (RT-PCR). Nude mice were inoculated with TE13 cells to establish ESCC xenografts. After two weeks, the inoculated mice were randomly divided into three groups. Tumors were injected with normal saline, transfection reagent pcDNA3.1(+) and transfection reagent pcDNA3.1(+)/RIZ1, respectively. Tumor development was quantified, and changes in gene expression of RIZ1 transfected tumors were detected by RT-PCR and Western blotting.
RESULTS: DNA microarray data showed that RIZ1 transfection induced widespread changes in gene expression profile of cell line TE13, with 960 genes upregulated and 1163 downregulated. Treatment of tumor xenografts with RIZ1 recombinant plasmid significantly inhibited tumor growth, decreased tumor size, and increased expression of RIZ1 mRNA compared to control groups. The changes in gene expression profile were also observed in vivo after RIZ1 transfection. Most of the differentially expressed genes were associated with cell development, supervision of viral replication, lymphocyte costimulatory and immune system development in esophageal cells. RIZ1 gene may be involved in multiple cancer pathways, such as cytokine receptor interaction and transforming growth factor beta signaling.
CONCLUSION: The development and progression of esophageal cancer are related to the inactivation of RIZ1. Virus infection may also be an important factor.
Keywords: Retinoblastoma protein-interacting zinc finger gene 1, Microarray, Nude mice, Esophageal squamous cell carcinoma cells
Core tip: Retinoblastoma protein-interacting zinc finger gene 1 (RIZ1) transfection induced widespread changes in gene expression profile of cell line TE13, with 960 genes upregulated and 1163 downregulated. Most of the differentially expressed genes are associated with cell development, supervision of viral replication, lymphocyte costimulatory, and immune system development in esophageal cells. RIZ1 gene may be involved in multiple cancer pathways, such as cytokine receptor interaction and transforming growth factor beta signaling. Virus infection may also be an important factor in the development of esophageal cancer.
INTRODUCTION
Esophageal cancer is one of the most common forms of cancer in the world and is a leading cause of cancer deaths in China and other developing countries. To date, the mechanisms of esophageal cancer are unclear. Tumor occurrence and development are regulated by a variety of oncogenes and tumor suppressor genes[1-3], including the putative tumor suppressor gene, Retinoblastoma protein-interacting zinc finger gene 1 (RIZ1). The RIZ gene has two expression products: RIZ1, which is believed to be a histone methyltransferase and acts on the locus of H3K9; and RIZ2, which lacks the PR-domain of RIZ1. Abnormal expression of RIZ1 has been found to be associated with tumor invasion and malignancy[4-10].
Our group has previously reported that RIZ1 expression level is lower in esophagus carcinoma than in adjacent noncancerous tissues[11], and is related to methylation of CpG islands[12]. In addition, by constructing human RIZ1 eukaryotic expression vectors to transfect human esophageal squamous cell carcinoma (ESCC) cell line TE13, we were able to report that upregulation of RIZ1 can recover tumor suppression activity and that treatment of cell line TE13 by methyltransferase inhibitor 5-aza-CdR reverses the methylation status of the promoter region[13]. In order to investigate RIZ1-mediated changes in gene expression of esophageal cancer, we compared the gene expression profile of TE13 cells transfected with RIZ1 with those of negative control cells. The resulting changes in oncogenicity were analyzed in vitro and by animal experimentation.
MATERIALS AND METHODS
Ethics
The animal study proposal was approved by the Tianjin Medical University General Hospital Ethics Committee with the permit number: 2012-021. All mouse experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China.
Cell culture and transfection
Human ESCC cell line TE13 was purchased from ATCC (Rcokville; MD, United States) and cultured in RPMI-1640 (HEPES 4.76 g/NaCO3 2.0 g/RPMI-1640 10.4 g/ddH2O 1000 mL) media supplemented with 10% new-born bovine serum, 2 mmol/L 1 × L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Life Technologies; NY, United States). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. RIZ1 eukaryotic expression vector pcDNA3.1(+)/RIZ1 plasmid had been prepared previously and stored at -80 °C. Cultivated TE13 cells were passaged in 12-inch orifice plates. Passaging was repeated every 2-3 d at 1:10 dilution, and cells were lifted by trypsin digestion. When the cells were at the log phase, they were transfected using the classical liposome method by adding 2 μL Lipofectamine 2000 (Life Technologies; NY, United States). Experimental and control groups were transfected with pcDNA3.1(+)/RIZ1 and pcDNA3.1(+), respectively. The media were changed after 6 h and the cells were washed in phosphate buffered saline (PBS), harvested and counted. After mixing with Trizol Reagent (Life Technologies, NY, United States), the cells were incubated at room temperature for 5 min, then transferred to liquid nitrogen. The resultant cDNA was taken and 0.75 μL was mixed with 2 × SYBR Premix Ex TaqTM (Takara). The following primer sets (10 μmol/L) were used: RIZ1, forward 5’-TCTGCTGTTGACAAGACCC-3’, reverse 5’-GCATCAATGCACATCCATC-3’; glyceraldehyde 3-phosphate dehydrogenase (GAPDH), forward 5’-ACCCAGAAGACTGTGGATGG-3’, reverse 5’-TTCAGCTCAGGGATGACCTT-3’. Amplification was carried out using LightCycler real-time polymerase chain reaction (PCR) system (Roche, United States), according to the manufacturer’s protocol. Each sample was run in triplicate for each gene. An initial denaturation step at 95 °C for 5 min was followed by 40 denaturation cycles at 94 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 30 s. A solubility temperature curve assay was constructed and the RIZ1 and GAPDH Ct-values for each group were recorded and compared. The RIZ1 mRNA relative quantitation formula: (2-∆∆Ct, × 100%), was applied to evaluate whether transfection was successful. Duplicate detections were performed in triplicate, through 2-∆∆Ct, to calculate mean ± SD.
Microarray analysis
Total RNA was extracted using Trizol Reagent (Life Technologies; NY, United States). Quality control was achieved by utilizing the Agilent Bioanalyzer 2100 (Agilent Technologies; United States). Purification was achieved using the RNeasy mini kit and RNase-Free DNase Set (QIAGEN, Germany). Agilent’s Low Input Quick Amp Labeling Kit, one-color and full genome chip (4 × 44K, design ID: 014850) were used to amplify and mark the mRNA according to the manufacturer’s protocols. The cRNA was purified and conjugated using the RNeasy mini kit. Agilent’s Gene Expression Hybridization Kit with a sample quantity of 1.65 μg cRNA was employed for gene chip hybridization for 17 h in a hybridization oven at < 65 °C and 10 rpm, following Agilent’s protocol. Slides were washed in staining dishes (Thermo Shandon, PA, United States) with Gene Expression Wash Buffer Kit. After completion of hybridization, gene chip scanning was performed using an Agilent Microarray Scanner. The software was adjusted to set the Dye channel to Green; scan resolution to 5 μm; and PMT to 100%, 10%, 16 bit. Data was read by Feature Extraction software v.10.7 and Gene Spring Software v.11.0, and uniformly processed by a Quantile algorithm. Data analysis was carried out using an online analysis system (SAS) (Shanghai Bohao Company, China). Fold changes ≥ 2 (upregulated) or ≤ 0.5 (downregulated) were set to select differentially expressed genes. Gene ontology (GO) functional analysis, chromosomal assignment, and pathway analysis were then performed. Several representative genes were selected for verification of the gene chip scan results by reverse transcription PCR (RT-PCR).
Animal experimentation
Purchase and feeding: Eighteen BALB/c nude mice (female; aged 4-6 wk; weight, 17-21 g), were purchased from the Experimental Animal Center of the Academy of Military Medical Sciences, Chinese People’s Liberation Army, under license No. SCXK-(army)-2007-004. Nude mice were fed in the Tianjin Medical University General Hospital SPF rearing chamber with sterilized standard feed and sterile water. Animal experimentation and protocol were carried out in accordance with medical ethical standards.
Subcutaneous transplantation: Cultured cells were adjusted to a final concentration of 1 × 107 cells/mL in sterilized PBS; 0.2 mL was injected into the right armpit of each nude mouse and their general condition and tumor growth after inoculation were observed. The large and small tumors in diameter were measured every three days, tumor size was calculated and a tumor growth curve plotted. The antitumor rate was calculated using the following formulae: Volume = 0.5 × small diameter × (large diameter)2; antitumor rate = (1 - the experimental group volume/the control group volume) × 100%.
Animal grouping and treatment: The nude mice were divided into three groups using a random digital method. Five mice from each group, with the maximum and minimum tumor volumes, were separated and the remaining mouse was removed. The blank control group was injected with 100 μL normal saline. The pcDNA3.1(+) group was injected with 20 μL liposome, to which 20 μg empty plasmid, adjusted to a final volume of 100 μL in DMEM, was added. The pcDNA3.1(+)/RIZ1 recombinant plasmid group was injected with 20 μL liposome to which 20 μg RIZ1 recombinant plasmid, adjusted to a final volume of 100 μL in DMEM, was added. Each group was injected every two days using the multi-point injection method, and the treatment was repeated 5 times. Seven days after the final injection, the mice were killed by cervical dislocation and the tumor tissue was removed, weighed and used for subsequent experimentation.
RT-PCR
RT-PCR was carried out as described above, and 12 mL of the reaction products were analyzed by electrophoresis on a 20 g/L agarose gel. The electrophoresis images were scanned by UV spectrophotometry (Beckman Coulter Inc., Brea, CA, United States).
Western blotting
Tissue from each group was homogenized in RIPA buffer (50 mmol/L Tris-HCl, pH 7.4; 150 mmol/L NaCl; 1% Nonidet P-40; 0.5% sodium deoxycholate; 0.1% SDS; 1 mmol/L EDTA; 1 mmol/L PMSF; 1 mg/mL Aprotinin). The supernatant was collected and protein concentrations were determined by a bicinchoninic acid (BCA) protein assay kit (Pierce; IL, United States); 30 μg of whole-cell lysate was separated on 8% SDS-PAGE gels, transferred to nitrocellulose (NC) membranes (Amersham Biosciences; New Jersey, United States), and immunoblotted with the following antibodies: anti-β-actin (ABCAM; United Kingdom), control; primary antibodies, 1:2000 dilution (ABCAM; United Kingdom); secondary antibodies Goat Anti-Mouse, 1:5000 dilution (ABCAM; United Kingdom). The films were analyzed by a PowerLook scanner (UMAX) and quantified by Image Quant software (GE; United States). The control experiments (TE13 cells; TE-13 cells transfected with blank plasmid) were treated by the same method. Relative expression of RIZ1 = gray value of RIZ1 protein/gray value of β-actin.
RESULTS
Transfection
The melting curve peaks for RIZ1 and GAPDH transcript products were at 80 °C and 82.5 °C, respectively (Figure 1), giving Ct-values for the two groups. The relative expression levels were calculated using the real-time PCR relative quantitative formula. RIZ1 gene expression levels were compared with SPSS v.13.0 statistical software. The results showed that the mRNA expression level in the experimental group was higher than in the control groups (Figure 2), indicating that transfection had been successful (P ≤ 0.01).
Figure 1.
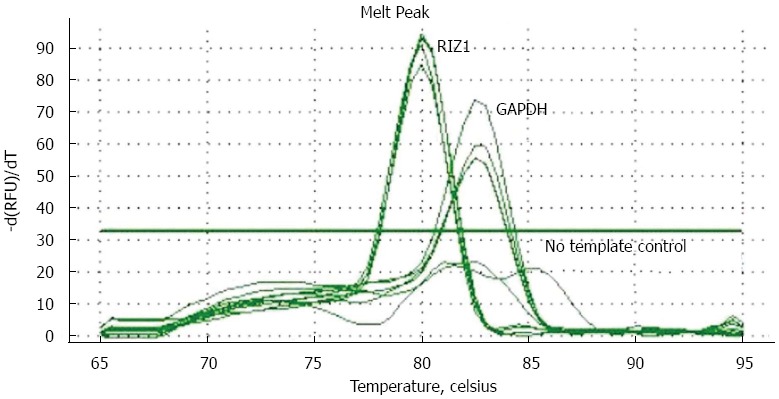
Solubility temperature curves for RIZ1 and glyceraldehyde 3-phosphate dehydrogenase showing that transfection was successful. DAPDH: Glyceraldehyde 3-phosphate dehydrogenase.
Figure 2.
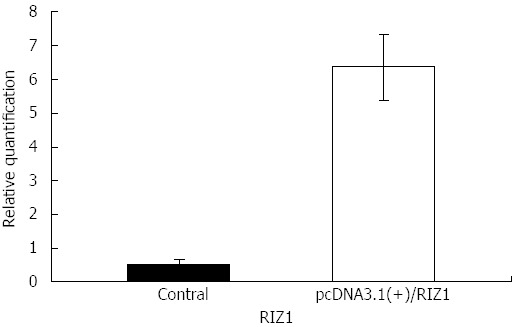
Histogram of RIZ1 gene mRNA expression levels in the blank control group and pcDNA3.1(+)/RIZ1 recombinant plasmid group, clearly showing that gene expression is higher in the pcDNA3.1(+)/RIZ1 group (P ≤ 0.01).
Alteration of gene expression profile
Table 1 gives the 2100 results for RIN ≥ 7.0 and 28S/18S ≥ 0.7, therefore qualifying the samples without degradation. The initial scanned single fluorescence chip data (Figure 3A) were standardized and converted to logarithmic values. A scatter plot was constructed with a two-dimensional rectangular coordinate plane (Figure 3B).
Table 1.
Sample qualification
| Sample number |
Concentration |
A260/A280 |
2100 result |
Results | |
| (μg/μL) | RIN | 28S/18S | |||
| 1 | 0.816 | 1.94 | 8.9 | 1.5 | Qualified |
| 2 | 0.865 | 1.93 | 8.9 | 1.5 | Qualified |
Figure 3.
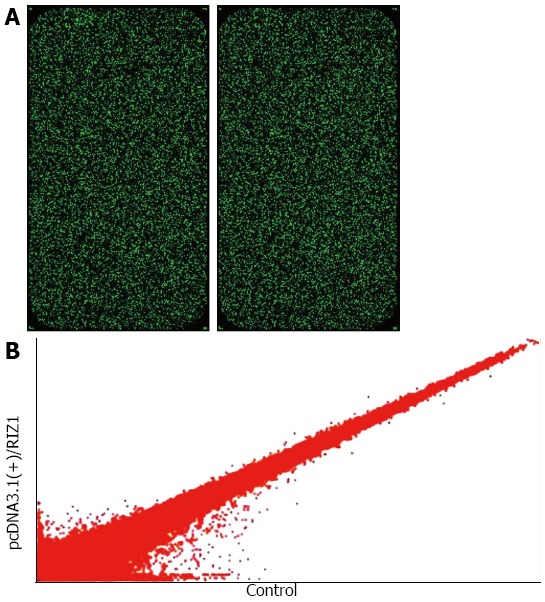
Alteration of gene expression profile. A: Single fluorescence chip images for microarray analysis; B: The data were used to construct the scatter plot. Hybridization signal strength scatter diagram. Each point in the scatter diagram represents a probe point on the chip. The position of each point is identified by an x and y coordinate: the x-coordinate gives the standardized signal value on the control chip; the y-coordinate gives the standardized signal value on the sample chip. The scatter plot is used to assess the centralized tendency of two sets of data.
GO and microarray analysis
The SAS system was used for GO analysis of the differentially expressed genes and P ≤ 0.05 was considered to be statistically significant (Table 2). The microarray data showed that 2123 genes were differentially expressed in the pcDNA3.1(+)/RIZ1 transfected cells with fold changes > 2 (P < 0.05) compared to control samples. Of these, 960 genes were upregulated, of which 654 were known genes (1.70%, 654/38500) and 306 were unknown; 1163 genes were downregulated, of which 719 were known genes (1.87%, 719/38500) and 444 were unknown. Subsequent analyses were primarily carried out on annotated genes. The gene chip results were confirmed by RT-PCR (Figure 4).
Table 2.
Gene ontology analysis of differentially expressed genes
| GO ID | Name | Hits | Total | Percentage | P value |
| GO: 0048468 | Cell development | 81 | 832 | 9.74% | 0.0288 |
| GO: 0050792 | Regulation of viral reproduction | 5 | 20 | 25.00% | 0.0304 |
| GO: 0031294 | Lymphocyte costimulation | 3 | 8 | 37.50% | 0.0397 |
| GO: 0002520 | Immune system development | 38 | 358 | 10.61% | 0.0428 |
Hits: Number of differentially expressed genes in the pathway; Total: The total number of genes in the pathway; Percentage: Percentage hits in the pathway (Hits/Total); P: Enrichment P value; GO: Gene ontology.
Figure 4.
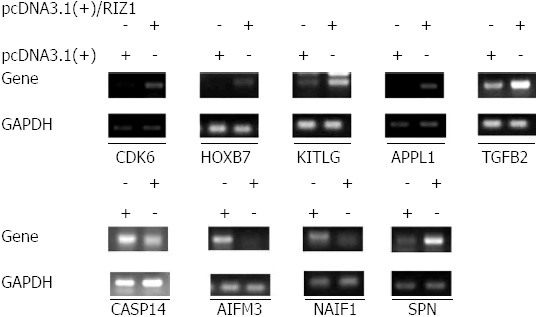
Comparison between results from the gene chip and the reverse transcription-polymerase chain reaction. Out of the nine genes tested, eight yielded consistent results for both reverse transcription-polymerase chain reaction and gene chip. The five upregulated genes are CDK6, HOXB7, KITLG, APPL1, TGFB2; and the three downregulated genes are CASP14, AIFM3 and NAIF1; however, in the case of service principal name (SPN), the reverse transcription-polymerase chain reaction results indicate upregulation, whereas the gene chip results indicate downregulation.
Pathway analysis
Many of the identified genes are associated with cell development, virus replication supervision, costimulatory molecule, and immune system development. Further analysis indicated that the RIZ1 gene may participate in multiple signaling pathways (P < 0.01), some of which are given in Table 3.
Table 3.
RIZ1 pathway analysis (selected pathways)
| Name | Hits | Total | Percentage | P value | q |
| Cytokine-cytokine receptor interaction | 25 | 276 | 9.06% | 0 | 0 |
| TGF-beta signaling pathway | 10 | 85 | 11.76% | 3.00E-04 | 5.00E-04 |
| MAPK signaling pathway | 20 | 271 | 7.38% | 2.00E-04 | 5.00E-04 |
| Pathways in cancer | 20 | 328 | 6.10% | 0.0018 | 0.0015 |
Hits: Number differentially expressed genes in the pathway; Total: Total number of genes in the pathway; Percentage: Percentage of hits in the pathway (Hits/Total); P: Enrichment P value; q: False discovery rate; TGF: Transforming growth factor; MAPK: Mitogen-activated protein kinase.
Transplantation and tumor growth
Transplantation of human esophageal cancer TE13 cells into nude mice was successful as shown in Figure 5. Tumor volumes were compared among the three groups (Figure 6A). The tumor growth rate curves revealed that tumor growth was slower in the pcDNA3.1(+)/RIZ1 group, with a shallower growth rate curve, than in the other two groups (Figure 6B). The following tumor volumes were recorded after 26 d: 1.025 ± 0.018 cm3 for the pcDNA3.1(+)/RIZ1 group, 2.208 ± 0.092 cm3 for the blank control group, and 1.980 ± 0.089 cm3 for the pcDNA3.1(+) group, showing that the tumor volume of the pcDNA3.1(+)/RIZ1 group was significantly lower than that of the other two groups (P < 0.05). In contrast, there was no significant difference between the blank control and pcDNA3.1(+) groups (P > 0.05).
Figure 5.
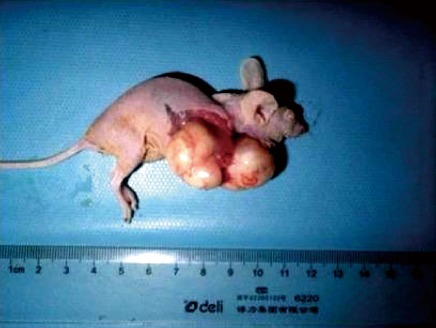
Photograph showing tumor development in a nude mouse, confirming successful transplantation of human esophageal cancer into an animal model.
Figure 6.
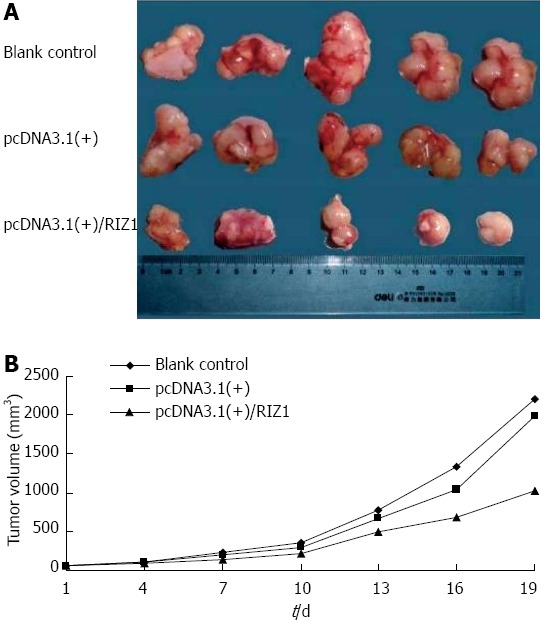
Transplantation and tumor growth. A: Photograph comparing esophageal squamous cell carcinomas removed from nude mice of the three groups after 26 d; B: Growth rate curves showing the increase in tumor volumes with time. The shallower curve in the pcDNA3.1(+)/RIZ1 group, compared to the blank control and pcDNA3.1(+) groups, indicates slower growth.
Electrophoresis
RNA was extracted from the transplanted tumors for RT-PCR, and the products were analyzed by electrophoresis: GAPDH was identified at 125 base pairs (bp) and RIZ1 at 167 bp. The intensity of the GAPDH band was similar among the three groups; however, the intensity of the RIZ1 band was greatest in the pcDNA3.1(+)/RIZ1 group, showing that RIZ1 expression level was higher than in the other two groups (Figure 7).
Figure 7.
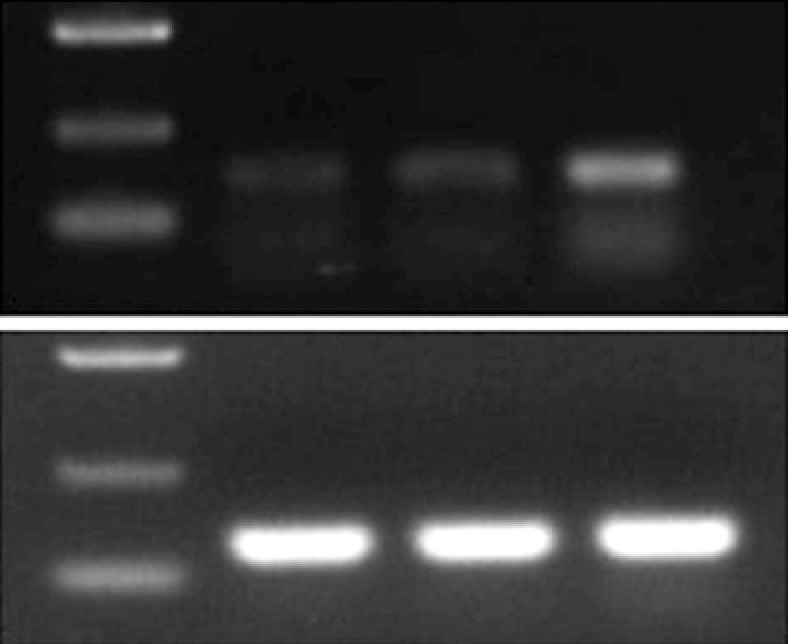
Electrophoresis gel image showing bands for glyceraldehyde 3-phosphate dehydrogenase (125 bp) and RIZI (167 bp). The intensity of the RIZ1 band is greatest in the pcDNA3.1(+)/RIZ1 group. M: Marker (D2000); N: Blank control; P: pcDNA3.1(+); R: pcDNA3.1(+)/RIZ1; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase.
Western blotting
The housekeeping gene, β-actin, was used as a control for Western blot analysis (Figure 8). The results showed no obvious difference in density of the β-actin bands among the three groups. In contrast, the density of the RIZ1 band was higher in the pcDNA3.1(+)/RIZ1 group than in the blank control and pcDNA3.1(+) groups, indicating that RIZ1 had been successfully expressed in the tumor tissues.
Figure 8.
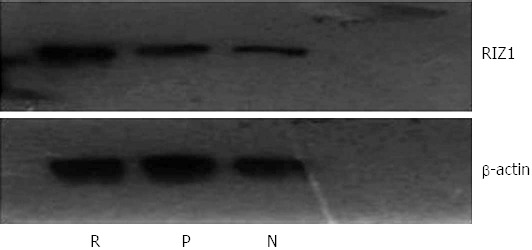
Western blot image for RIZ1 protein expression with β-actin as the control. The density of the RIZ1 band in the pcDNA3.1(+)/RIZ1 group is higher than in the other groups, indicating increased expression. R: pcDNA3.1(+)/RIZ1; N: Blank control; P: pcDNA3.1(+).
DISCUSSION
Esophageal cancer is the world’s most common cancer of the digestive system, with 70% of incidences occurring in China. China also has the highest incidence and mortality rates for both men and women. Recent statistics on morbidity and mortality for cancer patients show that esophageal cancer is the 6th most common form of cancer, and the 4th highest cause of cancer death-related in China[14-18]. Furthermore, the incidence rate is higher in rural areas than in urban areas. Esophageal cancer can be divided into two pathological types: ESCC and esophageal adenoid carcinoma. In China, ESCC accounts for 90% of esophageal cancers, in contrast to Western countries.
Treatment of esophageal carcinoma is a long-term project; however, by combining several different treatment types, the quality of life of the patients can be greatly improved. The purpose of our study is to enhance the understanding of ESCC development and mechanisms at the genetic level in order to advance clinical therapies. RIZ1 is one of the most effective tumor suppression genes, therefore failure of RIZ1 expression can lead to the development of many forms of cancer[19-25]. Our group has carried out a number of studies on the RIZ1 gene[11,12]; however, further research is required. One key research area is the introduction of a foreign gene into the tumor tissue, which is then able to express stably; eukaryotic expression vector is an ideal choice. The liposome mediated method was adopted because it has the advantages of high transfection efficiency, low immunogenicity, simple manipulation, and can be applied to a wide variety of cells.
We constructed the RIZ1 gene eukaryotic expression vector pcDNA3.1(+)/RIZ1 using an established molecular biology technique; empty plasmid, pcDNA3.1(+), was used as a negative control. After transfection into ESCC cell line TE13, in vivo experiments and gene chip analyses were carried out, as described in Materials and Methods. Our results showed that the xenograft in nude mice had a slower growth rate, and lower tumor volume and mass, in the pcDNA3.1(+)/RIZ1 group than in the blank control and pcDNA3.1(+) groups. This suggests that RIZ1 has a restraining effect on ESCC tumor growth. In contrast, the growth curves for the blank control and pcDNA3.1(+) groups were approximately parallel, indicating that these groups had no restraining effect. Secondly, after transfection, the gene expression profile of the cell line TE13 underwent extensive changes, with a total of 2123 differentially expressed genes, including 1163 downregulated and 960 upregulated. We found that many of these genes are involved in cell development, lymphocyte costimulatory, immune system development, and interestingly, in the supervision of viral replication. This is consistent with the results from Dąbrowski et al[13] who reported that low-risk type human papilloma virus may be one of the auxiliary activated or carcinogenic factors in ESCC occurrence and development. Vaiphei et al[26] also found that the infection rates of human papilloma virus in ESCC patients was as high as 87%, especially in patients with two or more types of phenotypic mixed infection. Persson et al[27] reported that HIV infection increases the risk of esophageal cancer. All these reports indicate that the occurrence and development of esophageal cancer may be related to virus infection; however, the pathophysiological mechanisms are poorly understood and require further research.
In conclusion, we propose that the occurrence of esophageal cancer is a consequence of widespread alterations in gene expression, involving multiple functions and signaling pathways with roles in tumor development, some of which are synergistic or antagonistic. We also speculate that the most probable signaling pathways in ESCC affected by these genes are the cytokine receptor interaction and transforming growth factor pathways. Therefore, we recommend that future research should be directed towards better understanding of the relationship between the RIZ1 gene and ESCC and the mechanism and role of virus infection in ESCC occurrence and development.
COMMENTS
Background
Esophageal cancer is one of the most common forms of cancer in the world and is a leading cause of cancer deaths in many developing countries. China is a country with a high incidence of esophageal cancer, and the pathological type is mainly the squamous cell carcinoma, which is different from the Western countries where adenocarcinoma is reported to be the main pathological type. To date, the mechanisms of esophageal cancer are unclear. Tumor occurrence and development are regulated by a variety of oncogenes and tumor suppressor genes, including the putative tumor suppressor gene, retinoblastoma protein-interacting zinc finger gene 1 (RIZ1).
Research frontiers
RIZ1 expression is lower in esophagus carcinoma and is related to methylation of CpG islands. In addition, by constructing human RIZ1 eukaryotic expression vectors to transfect human esophageal squamous cell carcinoma (ESCC) cell line TE13, the authors were able to report that upregulation of RIZ1 can recover tumor suppression activity and that treatment of cell line TE13 by methyltransferase inhibitor 5-aza-CdR reverses the methylation status of the promoter region.
Innovations and breakthroughs
In order to investigate RIZ1-mediated changes in gene expression of esophageal cancer, the authors compared the gene expression profile of TE13 cells transfected with RIZ1 with that of negative control cells. The resulting changes in oncogenicity were analyzed in vitro and by animal experimentation. They found that the development and progression of esophageal cancer are related with the inactivation of RIZ1. In addition, virus infection may also be an important factor.
Applications
RIZ1 is one of the most effective tumor suppression genes, therefore failure of RIZ1 expression can lead to the development of ESCC, which is expected to be a molecular biological parameter for early diagnosis.
Peer review
This paper reports that the development and progression of esophageal cancer is relevant to the inactivation of RIZ1. In addition, virus infection may also be an important factor.
Footnotes
Supported by The National Natural Science Foundation of China, No. 81201945; Science Foundation of Tianjin Medical University, No. 2011KY08; Doctoral Program of Higher Education Research Fund, No. 20091202110009; and Natural Science Foundation of Tianjin, China, No. 10JCYBJC11300
P- Reviewers Rajeshwari K, Rege RV S- Editor Gou SX L- Editor Ma JY E- Editor Ma S
References
- 1.Dong SW, Ma L, Xu N, Yan HQ, Liu HY, Li YW, Zhang P. Research on the reactivation of Syk expression caused by the inhibition of DNA promoter methylation in the lung cancer. Neoplasma. 2011;58:89–95. doi: 10.4149/neo_2011_01_89. [DOI] [PubMed] [Google Scholar]
- 2.Ma L, Dong S, Zhang P, Xu N, Yan H, Liu H, Li Y, Zhou Q. The relationship between methylation of the Syk gene in the promoter region and the genesis of lung cancer. Clin Lab. 2010;56:407–416. [PubMed] [Google Scholar]
- 3.Dong SW, Zhang P, Zhong RR, Liu Q. Expression of putative tumor suppressor gene spleen tyrosine kinase in esophageal squamous cell carcinoma. Clin Lab. 2013;59:647–653. doi: 10.7754/clin.lab.2012.120414. [DOI] [PubMed] [Google Scholar]
- 4.Buyse IM, Shao G, Huang S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc Natl Acad Sci USA. 1995;92:4467–4471. doi: 10.1073/pnas.92.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braig M, Schmitt CA. Oncogene-induced senescence: putting the brakes on tumor development. Cancer Res. 2006;66:2881–2884. doi: 10.1158/0008-5472.CAN-05-4006. [DOI] [PubMed] [Google Scholar]
- 6.Sasaki O, Meguro K, Tohmiya Y, Funato T, Shibahara S, Sasaki T. Altered expression of retinoblastoma protein-interacting zinc finger gene, RIZ, in human leukaemia. Br J Haematol. 2002;119:940–948. doi: 10.1046/j.1365-2141.2002.03972.x. [DOI] [PubMed] [Google Scholar]
- 7.Akahira J, Suzuki F, Suzuki T, Miura I, Kamogawa N, Miki Y, Ito K, Yaegashi N, Sasano H. Decreased expression of RIZ1 and its clinicopathological significance in epithelial ovarian carcinoma: correlation with epigenetic inactivation by aberrant DNA methylation. Pathol Int. 2007;57:725–733. doi: 10.1111/j.1440-1827.2007.02169.x. [DOI] [PubMed] [Google Scholar]
- 8.He L, Yu JX, Liu L, Buyse IM, Wang MS, Yang QC, Nakagawara A, Brodeur GM, Shi YE, Huang S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238–4244. [PubMed] [Google Scholar]
- 9.Chadwick RB, Jiang GL, Bennington GA, Yuan B, Johnson CK, Stevens MW, Niemann TH, Peltomaki P, Huang S, de la Chapelle A. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc Natl Acad Sci USA. 2000;97:2662–2667. doi: 10.1073/pnas.040579497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tam W, Gomez MF, Chadburn A, Knowles DM, Houldsworth J. Lack of A563G (I188V) missense mutation in RIZ/ PRDM2 in human diffuse large B-cell lymphomas. Genes Chromosomes Cancer. 2007;46:416–418. doi: 10.1002/gcc.20409. [DOI] [PubMed] [Google Scholar]
- 11.Dong SW, Cui YT, Zhong RR, Liang DC, Liu YM, Wang YG, He Z, Wang S, Liang SJ, Zhang P. Decreased expression of retinoblastoma protein-interacting zinc-finger gene 1 in human esophageal squamous cell cancer by DNA methylation. Clin Lab. 2012;58:41–51. [PubMed] [Google Scholar]
- 12.Dong SW, Zhang P, Liu YM, Cui YT, Wang S, Liang SJ, He Z, Sun P, Wang YG. Study on RIZ1 gene promoter methylation status in human esophageal squamous cell carcinoma. World J Gastroenterol. 2012;18:576–582. doi: 10.3748/wjg.v18.i6.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dąbrowski A, Kwaśniewski W, Skoczylas T, Bednarek W, Kuźma D, Goździcka-Józefiak A. Incidence of human papilloma virus in esophageal squamous cell carcinoma in patients from the Lublin region. World J Gastroenterol. 2012;18:5739–5744. doi: 10.3748/wjg.v18.i40.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 15.Chen BS, Wang MR, Xu X, Cai Y, Xu ZX, Han YL, Wu M. Transglutaminase-3, an esophageal cancer-related gene. Int J Cancer. 2000;88:862–865. doi: 10.1002/1097-0215(20001215)88:6<862::aid-ijc4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Metzger R, Schneider PM, Warnecke-Eberz U, Brabender J, Hölscher AH. Molecular biology of esophageal cancer. Onkologie. 2004;27:200–206. doi: 10.1159/000076913. [DOI] [PubMed] [Google Scholar]
- 17.Montesano R, Hollstein M, Hainaut P. Genetic alterations in esophageal cancer and their relevance to etiology and pathogenesis: a review. Int J Cancer. 1996;69:225–235. doi: 10.1002/(SICI)1097-0215(19960621)69:3<225::AID-IJC13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.Qin YR, Wang LD, Fan ZM, Kwong D, Guan XY. Comparative genomic hybridization analysis of genetic aberrations associated with development of esophageal squamous cell carcinoma in Henan, China. World J Gastroenterol. 2008;14:1828–1835. doi: 10.3748/wjg.14.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Shao G, Steele-Perkins G, Huang S. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J Biol Chem. 1997;272:2984–2991. doi: 10.1074/jbc.272.5.2984. [DOI] [PubMed] [Google Scholar]
- 20.Steele-Perkins G, Fang W, Yang XH, Van Gele M, Carling T, Gu J, Buyse IM, Fletcher JA, Liu J, Bronson R, et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 2001;15:2250–2262. doi: 10.1101/gad.870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa Y, Matsubara A, Teishima J, Seki M, Mita K, Usui T, Oue N, Yasui W. DNA methylation of the RIZ1 gene is associated with nuclear accumulation of p53 in prostate cancer. Cancer Sci. 2007;98:32–36. doi: 10.1111/j.1349-7006.2006.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lal G, Padmanabha L, Smith BJ, Nicholson RM, Howe JR, O’Dorisio MS, Domann FE. RIZ1 is epigenetically inactivated by promoter hypermethylation in thyroid carcinoma. Cancer. 2006;107:2752–2759. doi: 10.1002/cncr.22325. [DOI] [PubMed] [Google Scholar]
- 23.Gazzerro P, Abbondanza C, D’Arcangelo A, Rossi M, Medici N, Moncharmont B, Puca GA. Modulation of RIZ gene expression is associated to estradiol control of MCF-7 breast cancer cell proliferation. Exp Cell Res. 2006;312:340–349. doi: 10.1016/j.yexcr.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Tokumaru Y, Nomoto S, Jerónimo C, Henrique R, Harden S, Trink B, Sidransky D. Biallelic inactivation of the RIZ1 gene in human gastric cancer. Oncogene. 2003;22:6954–6958. doi: 10.1038/sj.onc.1206403. [DOI] [PubMed] [Google Scholar]
- 25.Fang W, Piao Z, Buyse IM, Simon D, Sheu JC, Perucho M, Huang S. Preferential loss of a polymorphic RIZ allele in human hepatocellular carcinoma. Br J Cancer. 2001;84:743–747. doi: 10.1054/bjoc.2000.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaiphei K, Kochhar R, Bhardawaj S, Dutta U, Singh K. High prevalence of human papillomavirus in esophageal squamous cell carcinoma: a study in paired samples. Dis Esophagus. 2013;26:282–287. doi: 10.1111/j.1442-2050.2012.01365.x. [DOI] [PubMed] [Google Scholar]
- 27.Persson EC, Shiels MS, Dawsey SM, Bhatia K, Anderson LA, Engels EA. Increased risk of stomach and esophageal malignancies in people with AIDS. Gastroenterology. 2012;143:943–950.e2. doi: 10.1053/j.gastro.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


