Fig. 2.2.
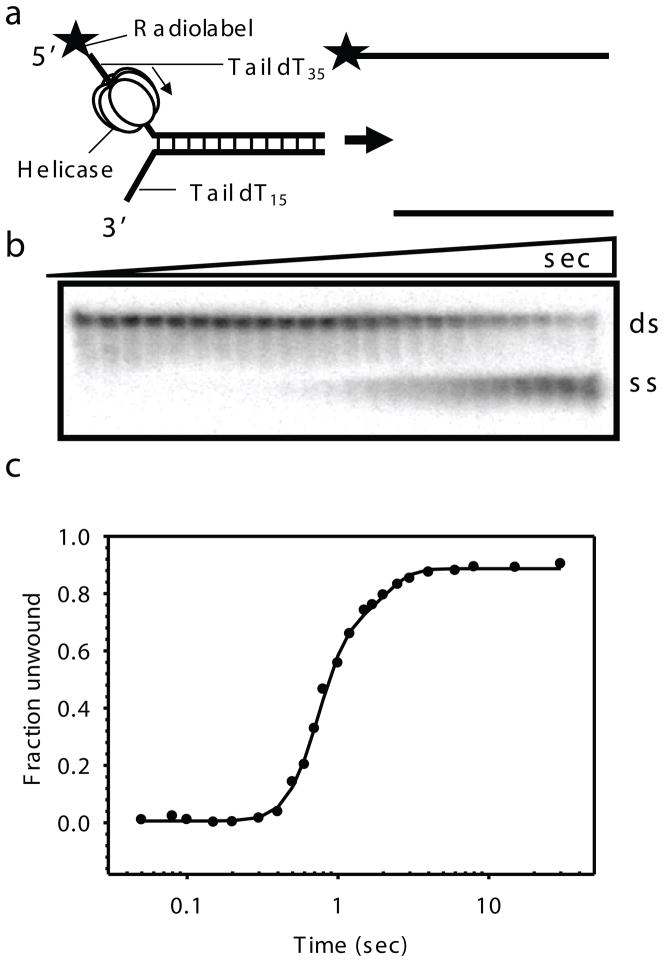
Gel-based radiometric assay for DNA unwinding. a) The DNA unwinding fork substrate design with radiolabeled top strand. T7 gp4 assembles on the top strand and moves in the 5′ to 3′ direction to unwind the dsDNA substrate. b) Representative native gel showing the ds and ss DNA resolved as a function of reaction time. Time points here are 0, 0.05, 0.08, 0.1, 0.15, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 1, 1.2, 1.5, 1.7, 2, 2.5, 3, 4, 6, 8, 15, 30 s (for 40ds duplex). c) Kinetics of ds40 unwinding at 18°C in reactions containing T7 gp4, dTTP, MgCl2 and SSB as the trap. The kinetics is fit to the gfit unwinding model (unwinding.m), which provides kf and s, from which the average rate of unwinding (kf × s) was determined. A good fit for the 40ds DNA unwinding data here gave parameters; A = 0.829, kf = 9.89, minD = 0, s = 6.34 and F0 = 0.002 and hence an average rate of unwinding ~ 62.7 bp/s.