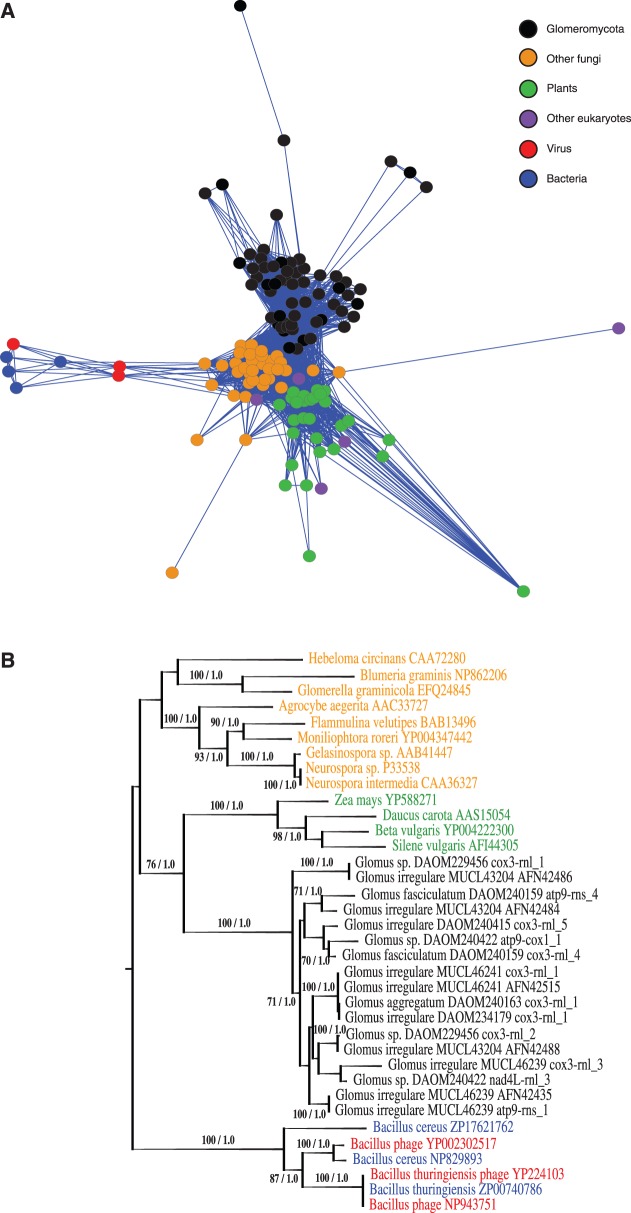
Fig. 4.—
(A) Sub-cluster of the global network of shared amino acid similarities between the Glomeromycota DPO proteins and all homologous sequences found on GenBank. Building this data set was performed using translated Glomeromycota sequences as queries for a BLASTp search on the GenBank nr database (minimal e-value threshold of 1E−40, 20% minimal similarity covering at least 20% of the smallest sequence). The network layouts were further produced by Cytoscape software, using an edge-weighted force-directed model, meaning that genes sharing more protein similarity appear closer in the display. There are 135 nodes in that network with 2,520 edges. (B) DPO protein maximum likelihood tree obtained with the rtREV + CAT phylogenetic model. The bacterial/viral cluster was used to root the tree. Number on branches indicates bootstrap support values (<60% cut-off) and Bayesian inference values, respectively. The Bayesian analyses gave a similar topology (data not shown). The tree includes AMF (black), other fungi (orange), plants (green), virus (red), and bacteria (blue).