Fig. 2.
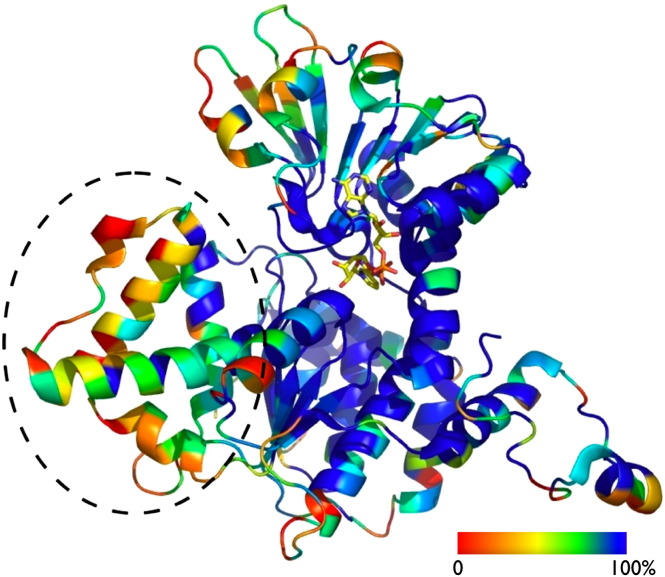
S-adenosyl-L-homocysteine hydrolase: sequence and structural conservation. The sequence conservation based on ConSurf calculations [160] using 166 unique S-adenosyl-L-homocysteine hydrolase sequences is color coded (from blue – identity to red – least conservation) and mapped onto the structure of the Pf-SAHH (PDB 1v8b [77]). Shown is one monomer of the functional tetrameric S-adenosyl-L-homocysteine hydrolase protein in cartoon drawing, the NAD+ cofactor (yellow) is shown in a stick representation and the substrate (Ado) was omitted for clarity. Note that the 40 amino acid insertion consisting of 3 helices and a loop (black circle), which is not present in mammalian and yeast S-adenosyl-L-homocysteine hydrolases, shows the least degree of conservation.