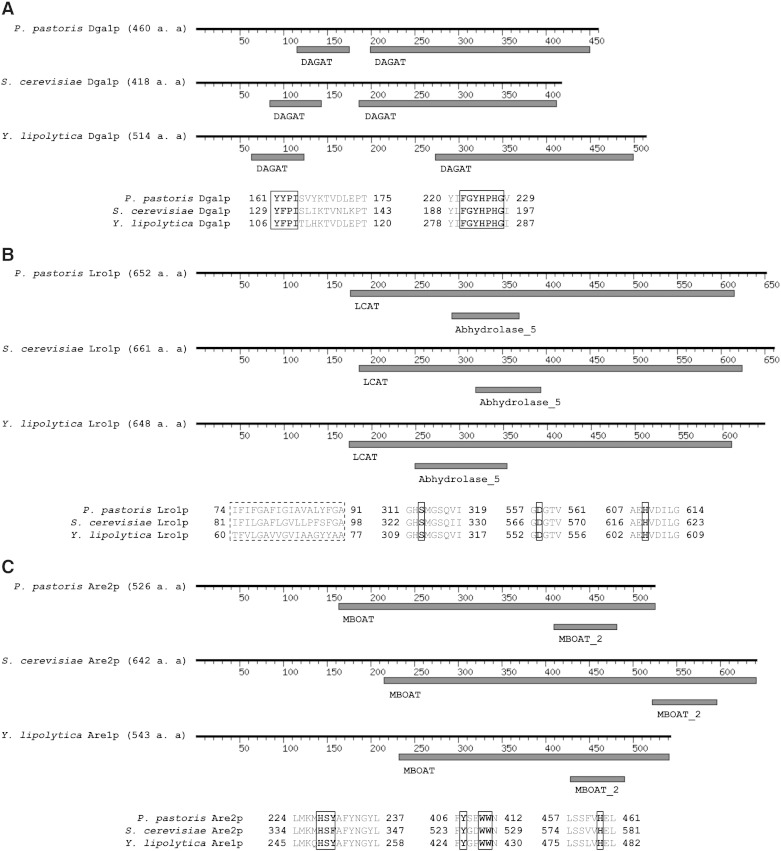
Fig. 1.
Sequence alignment of non-polar lipid synthesizing enzymes from P. pastoris.
A: Domain architecture of Dga1p from P. pastoris, S. cerevisiae and Y. lipolytica. Common diacylglycerol acyltransferase domains (DAGAT) are retrieved from Pfam database. Multiple protein sequence alignments show two highly conserved yeast Dga1p amino acid motifs (in bold, marked with squares). B: Domain architecture of Lro1p from P. pastoris, S. cerevisiae and Y. lipolytica. Common lecithin:cholesterol acyltransferase (LCAT) and α/β hydrolase (Abhydrolase_5) domains are retrieved from Pfam database. Aligned amino acid sequences of transmembrane domains are marked with dashed squares and active site vicinity motifs with potential active site catalytic triade (in bold) are shown within squares. C: Domain architecture of Are2p from P. pastoris, S. cerevisiae and Y. lipolytica. Common membrane bound O-acyl transferase family domains (MBOAT) are retrieved from Pfam database. Protein sequence alignment shows conserved active site motifs with crucial amino acids (in bold, marked with squares). Numbers indicate amino acid positions.