Figure 4.
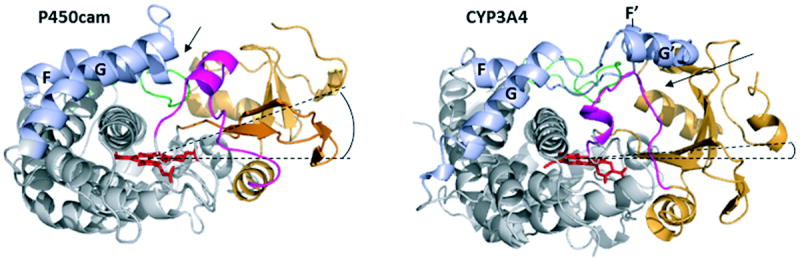
Comparison of the x-ray structures of soluble bacterial CYP101 (P450cam, PDB ID 1DZ4) and human CYP3A4 (1TQN). The beta-domain is depicted in orange, the B-B’ loop and B’ helix in magenta, the F, F’, G’ and G helices and connecting loops in light blue, the C-terminal loop in green, and the heme cofactor in red sticks. Dashed lines pass through the heme plane and the center of the beta-domain. The F’-G’ helix/loop insertion in membrane-bound mammalian CYPs shifts the beta-domain toward the heme plane36 (compare angles between the dashed lines), which opens a channel located between the B-B’ loop and the β1 and β3 sheets of the beta-domain (shown by an arrow). In contrast, in CYP101 and other soluble P450s substrates access the active site primarily through a channel formed by the F-G loop and B’ helix (indicated by an arrow).
