Figure 7.
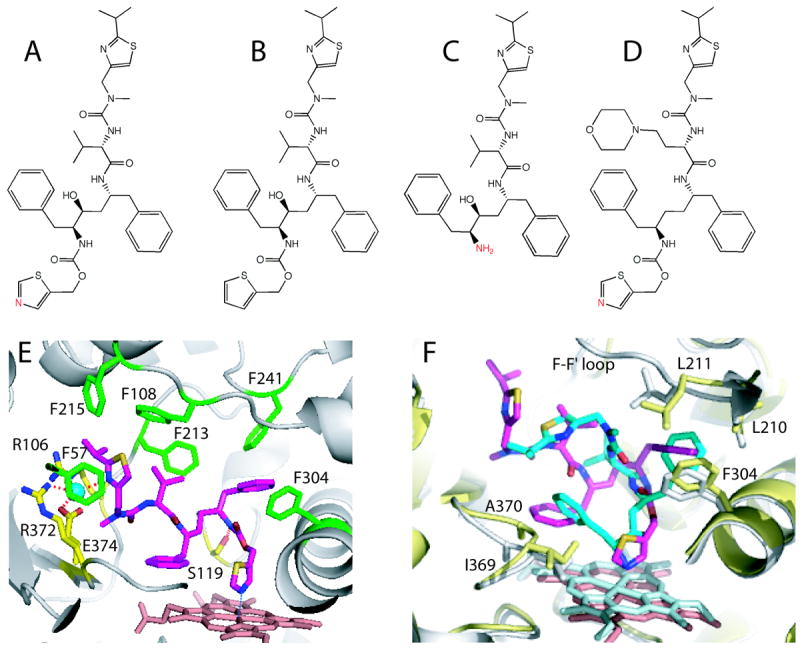
A-D, Chemical structures of ritonavir, deaza-ritonavir, DTMCR, and cobicistat, respectively. The heme-ligating primary amino group and thiazole nitrogens are shown in red. E, Ritonavir (magenta) bound to the active site of CYP3A4 (3NXU structure).54 Phenylalanine residues surrounding ritonavir are in green, whereas residues comprising the polar ‘umbrella’, a cluster of charged residues connected to the isopropyl moiety via a water molecule (cyan sphere), and the H-bond forming Ser119 are in yellow. F, Superposition of the ritonavir-(magenta, pink and gray) and DTMCR-bound (cyan, light cyan and yellow; 3TJS)33 structures of CYP3A4. To optimize hydrophobic interactions via phenyl side groups, DTMCR rotates by 180° relative to ritonavir. Since DTMCR is shorter than ritonavir, it cannot interact with the polar ‘umbrella’ and F-F’ loop. As a result, the F-F’ loop becomes disordered and the active site is solvent accessible in the DTMCR-bound structure. Also, DTMCR has differently oriented phenyl groups and does not clash with the 369-370 peptide. This is in contrast to the CYP3A4-ritonavir complex, where the heme shifts downwards and the Fe-N bond is slightly elongated because of steric hindrance with the 369-370 peptide.
